3F6O
 
 | | Crystal structure of ArsR family transcriptional regulator, RHA00566 | | Descriptor: | BROMIDE ION, Probable transcriptional regulator, ArsR family protein | | Authors: | Dong, A, Xu, X, Zheng, H, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ArsR family transcriptional regulator, RHA00566
To be Published
|
|
3F4A
 
 | | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family | | Descriptor: | AMMONIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Osipiuk, J, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family
To be Published
|
|
1XS7
 
 | | Crystal Structure of a cycloamide-urethane-derived novel inhibitor bound to human brain memapsin 2 (beta-secretase). | | Descriptor: | Beta-secretase 1, N-[(4S,5S,7R)-8-({(S)-1-[(BENZYLAMINO)OXOMETHYL]-2-METHYLPROPYL}AMINO)-5-HYDROXY-2,7-DIMETHYL-8-OXO-OCT-4-YL]-(4S,7S)-4 -ISOPROPYL-2,5,9-TRIOXO-1-OXA-3,6,10-TRIAZACYCLOHEXADECANE-7-CARBOXAMIDE | | Authors: | Ghosh, A, Devasamudram, T, Hong, L, DeZutter, C, Xu, X, Weerasena, V, Koelsch, G, Bilcer, G, Tang, J. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of cycloamide-urethane-derived novel inhibitors of human brain memapsin 2 (beta-secretase).
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3GEM
 
 | | Crystal structure of short-chain dehydrogenase from Pseudomonas syringae | | Descriptor: | ACETATE ION, Short chain dehydrogenase | | Authors: | Osipiuk, J, Xu, X, Cui, H, Nocek, B, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray crystal structure of short-chain dehydrogenase from Pseudomonas syringae.
To be Published
|
|
3GHD
 
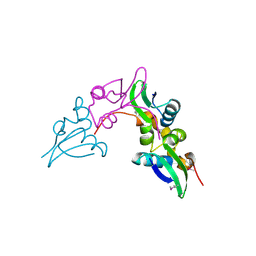 | | Crystal structure of a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain | | Descriptor: | a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain | | Authors: | Dong, A, Xu, X, Chruszcz, M, Brown, G, Proudfoot, M, Edwards, A.M, Joachimiak, A, Minor, W, Savchenko, A, Yaleunin, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain
To be Published
|
|
1XSV
 
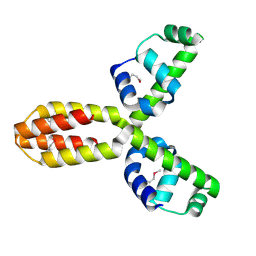 | | X-ray crystal structure of conserved hypothetical UPF0122 protein SAV1236 from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | Hypothetical UPF0122 protein SAV1236 | | Authors: | Walker, J.R, Xu, X, Virag, C, McDonald, M.-L, Houston, S, Buzadzija, K, Vedadi, M, Dharamsi, A, Fiebig, K.M, Savchenko, A. | | Deposit date: | 2004-10-20 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Crystal Structure of Conserved Hypothetical UPF0122 Protein SAV1236 From Staphylococcus aureus
To be Published
|
|
3K6H
 
 | | Crystal structure of a nitroreductase family protein from Agrobacterium tumefaciens str. C58 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase family protein, SULFATE ION | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of a nitroreductase family protein from Agrobacterium tumefaciens str. C58
To be Published
|
|
3CNI
 
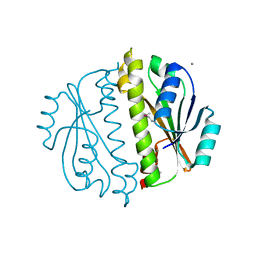 | | Crystal structure of a domain of a putative ABC type-2 transporter from Thermotoga maritima MSB8 | | Descriptor: | CALCIUM ION, Putative ABC type-2 transporter | | Authors: | Filippova, E.V, Shumilin, I, Tkaczuk, K.L, Cymborowski, M, Chruszcz, M, Xu, X, Que, Q, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of the putative ABC-type 2 transporter from Thermotoga maritima MSB8.
J.Struct.Funct.Genom., 15, 2014
|
|
3JU1
 
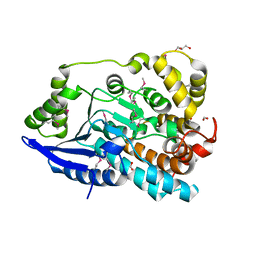 | | Crystal Structure of Enoyl-CoA Hydratase/Isomerase Family Protein | | Descriptor: | ACETIC ACID, Enoyl-CoA hydratase/isomerase family protein, FORMIC ACID, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal Structure of Enoyl-CoA Hydratase/Isomerase Family Protein
To be Published
|
|
3DNH
 
 | | The crystal structure of the protein Atu2129 (unknown function) from Agrobacterium tumefaciens str. C58 | | Descriptor: | uncharacterized protein Atu2129 | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-02 | | Release date: | 2008-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The crystal structure of the protein Atu2129 (unknown function) from Agrobacterium tumefaciens str. C58
To be Published
|
|
1Y8A
 
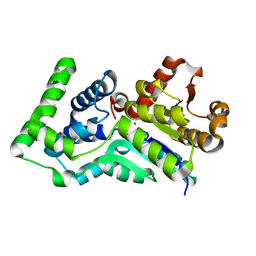 | | Structure of gene product AF1437 from Archaeoglobus fulgidus | | Descriptor: | MAGNESIUM ION, hypothetical protein AF1437 | | Authors: | Cuff, M.E, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of gene product AF1437 from Archaeoglobus fulgidus
To be published
|
|
3F5R
 
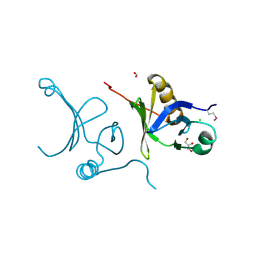 | | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p). | | Descriptor: | CHLORIDE ION, FACT complex subunit POB3, FORMIC ACID, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p).
To be Published
|
|
3EY7
 
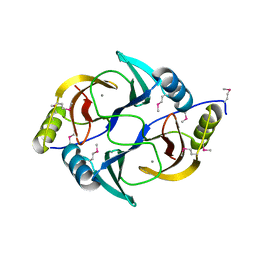 | | Structure from the mobile metagenome of V. Cholerae. Integron cassette protein VCH_CASS1 | | Descriptor: | Biphenyl-2,3-diol 1,2-dioxygenase III-related protein, CALCIUM ION | | Authors: | Harrop, S.J, Deshpande, C.N, Sureshan, V, Boucher, Y, Xu, X, Cui, H, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-20 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure from the mobile metagenome of V. Cholerae. Integron cassette protein VCH_CASS1
To be Published
|
|
3HCY
 
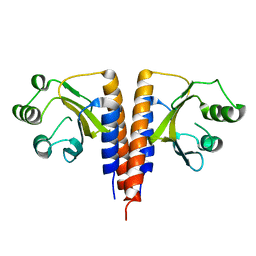 | | The crystal structure of the domain of putative two-component sensor histidine kinase protein from Sinorhizobium meliloti 1021 | | Descriptor: | Putative two-component sensor histidine kinase protein | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of the domain of putative two-component sensor histidine kinase protein from Sinorhizobium meliloti 1021
To be Published
|
|
3GL5
 
 | | Crystal structure of probable DsbA oxidoreductase SCO1869 from Streptomyces coelicolor | | Descriptor: | ACETATE ION, Putative DsbA oxidoreductase SCO1869, SODIUM ION | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of probable DsbA oxidoreductase SCO1869 from Streptomyces coelicolor
To be Published
|
|
3EXM
 
 | | Crystal structure of the phosphatase SC4828 with the non-hydrolyzable nucleotide GPCP | | Descriptor: | CALCIUM ION, GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID GUANOSYL ESTER, ... | | Authors: | Singer, A.U, Xu, X, Zheng, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-16 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of a new family of prokaryotic nucleoside diphosphatases.
To be Published
|
|
3DTZ
 
 | | Crystal structure of Putative Chlorite dismutase TA0507 | | Descriptor: | FORMIC ACID, Putative Chlorite dismutase TA0507 | | Authors: | Chang, C, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-16 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of Putative Chlorite dismutase TA0507
To be Published
|
|
3HFI
 
 | | The crystal structure of the putative regulator from Escherichia coli CFT073 | | Descriptor: | Putative regulator | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-11 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the putative regulator from Escherichia coli CFT073
To be Published
|
|
3FHM
 
 | | Crystal structure of the CBS-domain containing protein ATU1752 from Agrobacterium tumefaciens | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Zhang, R, Cui, H, Kudritsdka, M, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the CBS-domain containing protein ATU1752 from Agrobacterium tumefaciens
To be Published
|
|
1YX1
 
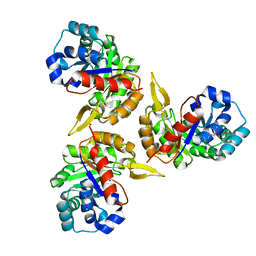 | | Crystal Structure of Protein of Unknown Function PA2260 from Pseudomonas aeruginosa, Possible Sugar Phosphate Isomerase | | Descriptor: | ISOPROPYL ALCOHOL, SODIUM ION, hypothetical protein PA2260 | | Authors: | Osipiuk, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-19 | | Release date: | 2005-04-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of hypothetical protein PA2260 from Pseudomonas aeruginosa
To be Published
|
|
3K0B
 
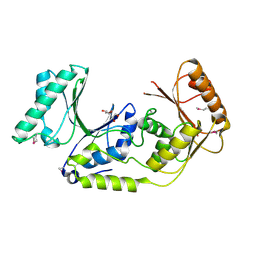 | | Crystal structure of a predicted N6-adenine-specific DNA methylase from Listeria monocytogenes str. 4b F2365 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, predicted N6-adenine-specific DNA methylase | | Authors: | Nocek, B, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-24 | | Release date: | 2009-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a predicted N6-adenine-specific DNA methylase from Listeria monocytogenes str. 4b F2365
To be Published
|
|
1Z1S
 
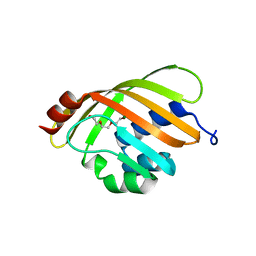 | | Crystal Structure of Putative Isomerase PA3332 from Pseudomonas aeruginosa | | Descriptor: | Hypothetical Protein PA3332, MAGNESIUM ION, TRIETHYLENE GLYCOL | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-06 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein PA3332 from Pseudomonas aeruginosa
To be Published
|
|
3F6V
 
 | | Crystal structure of Possible transcriptional regulator for arsenical resistance | | Descriptor: | MAGNESIUM ION, Possible transcriptional regulator, ArsR family protein | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of Possible transcriptional regulator for arsenical resistance from Rhodococcus sp.
To be Published
|
|
3FBU
 
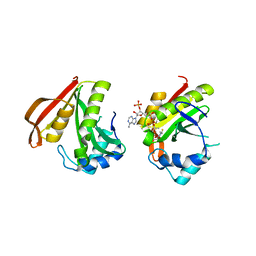 | | The crystal structure of the acetyltransferase (GNAT family) from Bacillus anthracis | | Descriptor: | Acetyltransferase, GNAT family, COENZYME A | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the acetyltransferase (GNAT family) from Bacillus anthracis
To be Published
|
|
3FM5
 
 | | X-ray crystal structure of transcriptional regulator (MarR family) from Rhodococcus sp. RHA1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nocek, B, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of transcriptional regulator (MarR family) from Rhodococcus sp. RHA1
To be Published
|
|
