8IOY
 
 | | Structure of ATP7B C983S/C985S/D1027A mutant with AMP-PNP | | Descriptor: | Copper-transporting ATPase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yang, G, Xu, L, Guo, J, Wu, Z. | | Deposit date: | 2023-03-13 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
6A96
 
 | | Cryo-EM structure of the human alpha5beta3 GABAA receptor in complex with GABA and Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-5,Gamma-aminobutyric acid receptor subunit alpha-5, ... | | Authors: | Liu, S, Xu, L, Guan, F, Liu, Y.T, Cui, Y, Zhang, Q, Bi, G.Q, Zhou, Z.H, Zhang, X, Ye, S. | | Deposit date: | 2018-07-11 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM structure of the human alpha 5 beta 3 GABAAreceptor.
Cell Res., 28, 2018
|
|
7XUN
 
 | |
7XUO
 
 | | Structure of ATP7B C983S/C985S/D1027A mutant with cisplatin in presence of ATOX1 | | Descriptor: | Copper-transporting ATPase 2, PLATINUM (II) ION | | Authors: | Yang, G, Xu, L, Chang, S, Guo, J, Wu, Z. | | Deposit date: | 2022-05-19 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
7XUK
 
 | |
7XUM
 
 | | Structure of ATP7B C983S/C985S/D1027A mutant with Cu+ in presence of ATOX1 | | Descriptor: | COPPER (II) ION, Copper-transporting ATPase 2 | | Authors: | Yang, G, Xu, L, Chang, S, Guo, J, Wu, Z. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
5WHZ
 
 | | PGDM1400-10E8v4 CODV Fab | | Descriptor: | Anti-HIV CODV-Fab Heavy chain, Anti-HIV CODV-Fab Light chain | | Authors: | Lord, D.M, Wei, R.R. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.549 Å) | | Cite: | Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques.
Science, 358, 2017
|
|
5XS5
 
 | | Structure of Coxsackievirus A6 (CVA6) virus procapsid particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Cheng, T, Li, S.W. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5XS7
 
 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle in complex with the neutralizing antibody fragment 1D5 | | Descriptor: | Genome polyprotein, Heavy chain of Fab 1D5, Light chain of Fab 1D5 | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5XS4
 
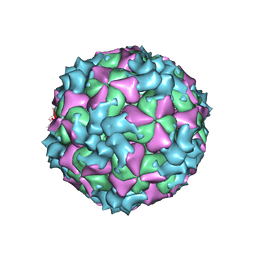 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
7VS7
 
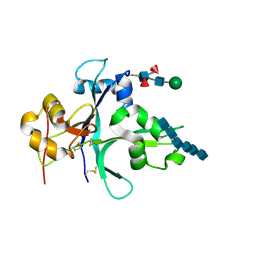 | | Crystal structure of the ectodomain of OsCERK1 in complex with chitin hexamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Li, X. | | Deposit date: | 2021-10-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structural insight into chitin perception by chitin elicitor receptor kinase 1 of Oryza sativa.
J Integr Plant Biol, 65, 2023
|
|
7DPF
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion | | Descriptor: | Capsid protein VP4, PALMITIC ACID, VP2, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DF5
 
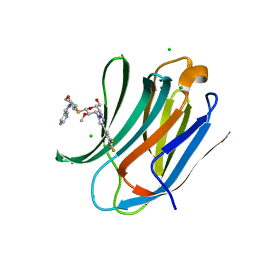 | | Human Galectin-3 CRD in complex with novel tetrahydropyran-based thiodisaccharide mimic inhibitor | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-5-methoxy-6-[(3R,4R,5S)-4-oxidanyl-5-(4-pyrimidin-5-yl-1,2,3-triazol-1-yl)oxan-3-yl]sulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-3-ol, CHLORIDE ION, Galectin-3, ... | | Authors: | Ghosh, K, Kumar, A. | | Deposit date: | 2020-11-06 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Synthesis, Structure-Activity Relationships, and In Vivo Evaluation of Novel Tetrahydropyran-Based Thiodisaccharide Mimics as Galectin-3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7DF6
 
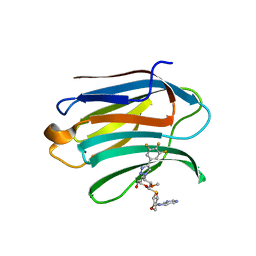 | | Mouse Galectin-3 CRD in complex with novel tetrahydropyran-based thiodisaccharide mimic inhibitor | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-5-methoxy-6-[(3R,4R,5S)-4-oxidanyl-5-(4-pyrimidin-5-yl-1,2,3-triazol-1-yl)oxan-3-yl]sulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-3-ol, Galectin-3 | | Authors: | Ghosh, K, Kumar, A. | | Deposit date: | 2020-11-06 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, Structure-Activity Relationships, and In Vivo Evaluation of Novel Tetrahydropyran-Based Thiodisaccharide Mimics as Galectin-3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
8GXP
 
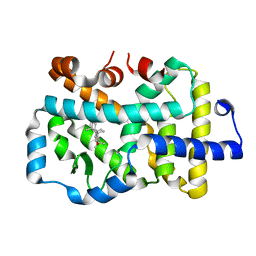 | | Complex structure of RORgama with betulinic acid | | Descriptor: | Betulinic acid, Nuclear receptor ROR-gamma | | Authors: | Zhang, X.L, Xu, C, Bai, F. | | Deposit date: | 2022-09-20 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery, structural optimization, and anti-tumor bioactivity evaluations of betulinic acid derivatives as a new type of ROR gamma antagonists.
Eur.J.Med.Chem., 257, 2023
|
|
4DBH
 
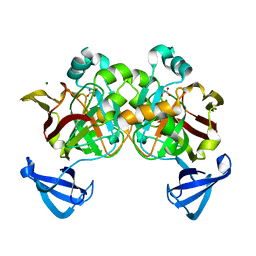 | | Crystal structure of Cg1458 with inhibitor | | Descriptor: | 2-HYDROXYHEPTA-2,4-DIENE-1,7-DIOATE ISOMERASE, MAGNESIUM ION, OXALATE ION | | Authors: | Ran, T.T, Wang, W.W, Xu, D.Q, Gao, Y.Y. | | Deposit date: | 2012-01-15 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of Cg1458 reveal a catalytic lid domain and a common catalytic mechanism for FAH family.
Biochem.J., 449, 2013
|
|
4DBF
 
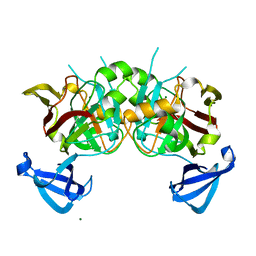 | | Crystal structures of Cg1458 | | Descriptor: | 2-HYDROXYHEPTA-2,4-DIENE-1,7-DIOATE ISOMERASE, MAGNESIUM ION | | Authors: | Ran, T.T, Xu, D.Q, Wang, W.W, Gao, Y.Y, Wang, M.T. | | Deposit date: | 2012-01-15 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Cg1458 reveal a catalytic lid domain and a common catalytic mechanism for FAH family.
Biochem.J., 449, 2013
|
|
4WNM
 
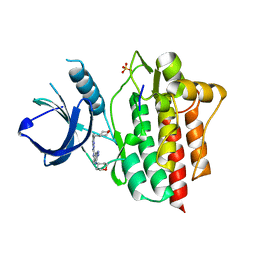 | | SYK catalytic domain in complex with a potent triazolopyridine inhibitor | | Descriptor: | N~3~-(tetrahydro-2H-pyran-4-yl)-N~6~-[5-(tetrahydro-2H-pyran-4-ylmethyl)[1,2,4]triazolo[1,5-a]pyridin-2-yl]-1H-indazole-3,6-diamine, SULFATE ION, Tyrosine-protein kinase SYK | | Authors: | Jackson, P.J. | | Deposit date: | 2014-10-13 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Triazolopyridine-Based Spleen Tyrosine Kinase Inhibitor That Arrests Joint Inflammation.
Plos One, 11, 2016
|
|
4Y7N
 
 | | The Structure Insight into 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
4Y52
 
 | | Crystal structure of 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-11 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
8JHB
 
 | | FZD6 Gs complex | | Descriptor: | Frizzled-6, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Zhang, Z. | | Deposit date: | 2023-05-23 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8J9O
 
 | | Cryo-EM structure of inactive FZD1 | | Descriptor: | Frizzled-1,Soluble cytochrome b562, anti-BRIL Fab Heavy Chain, anti-BRIL Fab Light Chain, ... | | Authors: | Lin, X, Xu, F. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8JH7
 
 | | FZD6 in inactive state | | Descriptor: | Frizzled-6,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain, ... | | Authors: | Xu, F, Zhang, Z. | | Deposit date: | 2023-05-22 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8JHC
 
 | | FZD3 in inactive state | | Descriptor: | Frizzled-3,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain, ... | | Authors: | Xu, F, Zhang, Z. | | Deposit date: | 2023-05-23 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8JHI
 
 | | FZD3-Gs complex | | Descriptor: | Frizzled-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Zhang, Z. | | Deposit date: | 2023-05-23 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
