7S5H
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 35 | | Descriptor: | (2E)-but-2-ene-1,4-diol, Pro-peptide from Proprotein convertase subtilisin/kexin type 9, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Orth, P. | | Deposit date: | 2021-09-10 | | Release date: | 2021-11-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.272 Å) | | Cite: | A Series of Novel, Highly Potent, and Orally Bioavailable Next-Generation Tricyclic Peptide PCSK9 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7S5G
 
 | | PCSK9 in complex with compound 19 | | Descriptor: | (2E)-but-2-ene-1,4-diol, GLYCEROL, Propeptide of Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Orth, P. | | Deposit date: | 2021-09-10 | | Release date: | 2021-11-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | A Series of Novel, Highly Potent, and Orally Bioavailable Next-Generation Tricyclic Peptide PCSK9 Inhibitors.
J.Med.Chem., 64, 2021
|
|
3GV3
 
 | |
7N61
 
 | | structure of C2 projections and MIPs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FAP147, FAP178, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-07 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7N6G
 
 | | C1 of central pair | | Descriptor: | CPC1, Calmodulin, DPY30, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-08 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3PUR
 
 | | CEKDM7A from C.Elegans, complex with D-2-HG | | Descriptor: | (2R)-2-hydroxypentanedioic acid, FE (II) ION, Lysine-specific demethylase 7 homolog, ... | | Authors: | Yang, Y, Wang, P, Xu, W, Xu, Y. | | Deposit date: | 2010-12-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases
Cancer Cell, 19, 2011
|
|
3PUQ
 
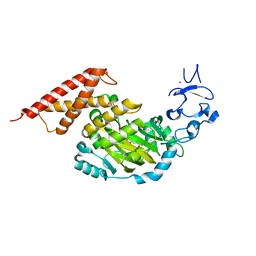 | | CEKDM7A from C.Elegans, complex with alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, GLYCEROL, ... | | Authors: | Yang, Y, Wang, P, Xu, W, Xu, Y. | | Deposit date: | 2010-12-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases
Cancer Cell, 19, 2011
|
|
8Q7E
 
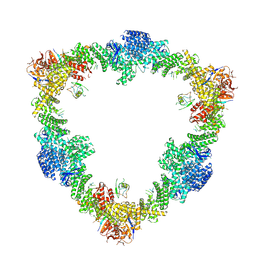 | |
8Q7H
 
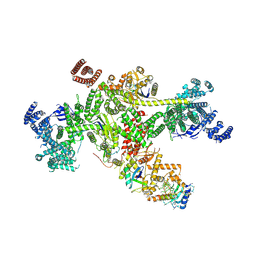 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, NEDD8, ... | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Schulman, B.A. | | Deposit date: | 2023-08-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 2024
|
|
8RHZ
 
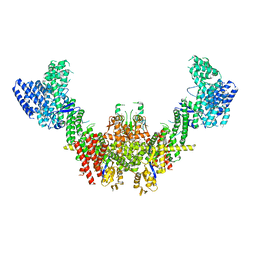 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated conformation - symmetry expanded unneddylated dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2023-12-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 2024
|
|
4WF6
 
 | | Anthrax toxin lethal factor with bound small molecule inhibitor MK-31 | | Descriptor: | 1,2-ETHANEDIOL, Lethal factor, N~2~-[(4-fluoro-3-methylphenyl)sulfonyl]-N-hydroxy-D-alaninamide, ... | | Authors: | Maize, K.M, De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2014-09-12 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6521 Å) | | Cite: | Probing the S2' Subsite of the Anthrax Toxin Lethal Factor Using Novel N-Alkylated Hydroxamates.
J.Med.Chem., 58, 2015
|
|
5APH
 
 | | Ligand complex of RORg LBD | | Descriptor: | DIMETHYL SULFOXIDE, N-(2-FLUOROPHENYL)-4-[(4-FLUOROPHENYL)SULFONYL]-2,3,4,5-TETRAHYDRO-1,4-BENZOXAZEPIN-6-AMINE, NUCLEAR RECEPTOR COACTIVATOR 2, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2015-09-16 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Benzoxazepines Achieve Potent Suppression of IL-17 Release in Human T-Helper 17 (TH 17) Cells through an Induced-Fit Binding Mode to the Nuclear Receptor ROR gamma.
ChemMedChem, 11, 2016
|
|
5APK
 
 | | Ligand complex of RORg LBD | | Descriptor: | 2-CHLORO-6-FLUORO-N-[4-[3-(TRIFLUOROMETHYL)PHENYL]SULFONYL-3,5-DIHYDRO-2H-1,4-BENZOXAZEPIN-7-YL]BENZAMIDE, NUCLEAR RECEPTOR ROR-GAMMA | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2015-09-16 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Benzoxazepines Achieve Potent Suppression of IL-17 Release in Human T-Helper 17 (TH 17) Cells through an Induced-Fit Binding Mode to the Nuclear Receptor ROR gamma.
ChemMedChem, 11, 2016
|
|
5APJ
 
 | | Ligand complex of RORg LBD | | Descriptor: | 2-CHLORO-6-FLUORO-N-[4-[3-(TRIFLUOROMETHYL)PHENYL]SULFONYL-3,5-DIHYDRO-2H-1,4-BENZOXAZEPIN-7-YL]BENZAMIDE, NUCLEAR RECEPTOR COACTIVATOR 2, NUCLEAR RECEPTOR ROR-GAMMA, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2015-09-16 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Benzoxazepines Achieve Potent Suppression of IL-17 Release in Human T-Helper 17 (TH 17) Cells through an Induced-Fit Binding Mode to the Nuclear Receptor ROR gamma.
ChemMedChem, 11, 2016
|
|
6U3U
 
 | | Crystal Structure of Shiga Toxin 2K | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Shiga toxin 2K subunit A, ... | | Authors: | Zhang, Y.Z, He, X.H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Structural and Functional Characterization of Stx2k, a New Subtype of Shiga Toxin 2.
Microorganisms, 8, 2019
|
|
7VWY
 
 | |
7VWZ
 
 | | Cryo-EM structure of Rob-dependent transcription activation complex in a unique conformation | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-11-12 | | Release date: | 2022-06-08 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription activation by Rob, a pleiotropic AraC/XylS family regulator.
Nucleic Acids Res., 50, 2022
|
|
4FFW
 
 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with Fab + sitagliptin | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, Dipeptidyl peptidase 4, Fab heavy chain, ... | | Authors: | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | Deposit date: | 2012-06-01 | | Release date: | 2012-12-12 | | Last modified: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
4FFV
 
 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with 11A19 Fab | | Descriptor: | 11A19 Fab heavy chain, 11A19 Fab light chain, Dipeptidyl peptidase 4 | | Authors: | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | Deposit date: | 2012-06-01 | | Release date: | 2012-12-12 | | Last modified: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
8CJ7
 
 | | HDAC6 selective degraded (difluoromethyl)-1,3,4-oxadiazole substrate inhibitor | | Descriptor: | 6-[(5-pyridin-2-yl-1,2$l^{4},3,4-tetrazacyclopenta-1,3-dien-2-yl)methyl]pyridine-3-carbohydrazide, Histone deacetylase 6, IODIDE ION, ... | | Authors: | Sandmark, J, Ek, M, Ripa, L. | | Deposit date: | 2023-02-12 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Selective and Bioavailable HDAC6 2-(Difluoromethyl)-1,3,4-oxadiazole Substrate Inhibitors and Modeling of Their Bioactivation Mechanism.
J.Med.Chem., 66, 2023
|
|
5XHQ
 
 | | Apolipoprotein N-acyl Transferase | | Descriptor: | Apolipoprotein N-acyltransferase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yingzhi, X, Yong, X, Guangyuan, L, Fei, S. | | Deposit date: | 2017-04-23 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Crystal structure of E. coli apolipoprotein N-acyl transferase
Nat Commun, 8, 2017
|
|
6V3Z
 
 | |
6U2N
 
 | | PCSK9 in complex with compound 4 | | Descriptor: | 4-{[(1R)-6-methoxy-1-methyl-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}benzoic acid, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-20 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
6U36
 
 | | PCSK9 in complex with a Fab and compound 14 | | Descriptor: | 2-fluoro-4-{[(1R)-6-(2-{4-[1-(4-methoxyphenyl)-5-methyl-6-oxo-1,6-dihydropyridazin-3-yl]-1H-1,2,3-triazol-1-yl}ethoxy)-1-methyl-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}benzoic acid, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-21 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
6U3X
 
 | | PCSK9 in complex with compound 2 | | Descriptor: | 2-[(1R)-6,7-dimethoxy-1-methyl-1,2,3,4-tetrahydroisoquinolin-1-yl]-N-(1,3-thiazol-2-yl)acetamide, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-22 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
