6J1S
 
 | | Crystal structure of Candida Antarctica Lipase B mutant - SS | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Lipase B, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
6J1T
 
 | | Crystal structure of Candida Antarctica Lipase B mutant SR with product analogue | | Descriptor: | (2S)-2-phenyl-N-[(1R)-1-phenylethyl]propanamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
6J1P
 
 | | Crystal structure of Candida Antarctica Lipase B mutant - SR | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
6J1R
 
 | | Crystal structure of Candida Antarctica Lipase B mutant - RR | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Lipase B, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
7XVE
 
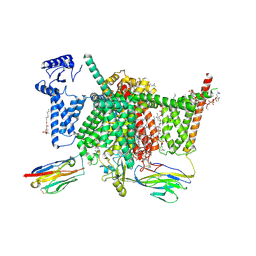 | | Human Nav1.7 mutant class-I | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-21 | | Release date: | 2022-08-10 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XVF
 
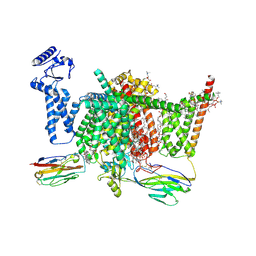 | | Nav1.7 mutant class2 | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-22 | | Release date: | 2022-08-10 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6ISQ
 
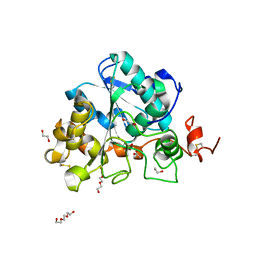 | | structure of Lipase mutant with oxided Cys-His-Asp catalytic triad | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-11-18 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Artificial cysteine-lipases with high activity and altered catalytic mechanism created by laboratory evolution.
Nat Commun, 10, 2019
|
|
6ISR
 
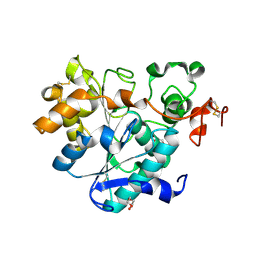 | | structure of lipase mutant with Cys-His-Asp catalytic triad | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lipase B, NICKEL (II) ION, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-11-18 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Artificial cysteine-lipases with high activity and altered catalytic mechanism created by laboratory evolution.
Nat Commun, 10, 2019
|
|
6ISP
 
 | | structure of Candida antarctica Lipase B mutant | | Descriptor: | CALCIUM ION, Lipase B, N,N-BIS(3-D-GLUCONAMIDOPROPYL)DEOXYCHOLAMIDE | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-11-18 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Artificial cysteine-lipases with high activity and altered catalytic mechanism created by laboratory evolution.
Nat Commun, 10, 2019
|
|
8BVL
 
 | |
8I5X
 
 | | Structure of human Nav1.7 in complex with Vinpocetine | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5B
 
 | | Structure of human Nav1.7 in complex with bupivacaine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5G
 
 | | Structure of human Nav1.7 in complex with PF-05089771 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5Y
 
 | | Structure of human Nav1.7 in complex with vixotrigine | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8J4F
 
 | | Structure of human Nav1.7 in complex with Hardwickii acid | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (4~{a}~{R},5~{S},6~{R},8~{a}~{R})-5-[2-(furan-3-yl)ethyl]-5,6,8~{a}-trimethyl-3,4,4~{a},6,7,8-hexahydronaphthalene-1-carboxylic acid, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-04-19 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8S9C
 
 | | Cryo-EM structure of Nav1.7 with CBZ | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8S9B
 
 | | Cryo-EM structure of Nav1.7 with LCM | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
5JLY
 
 | | Structure of Peroxiredoxin-1 from Schistosoma japonicum | | Descriptor: | Thioredoxin peroxidase-1 | | Authors: | Wu, Q, Huang, F, Zeng, D, Liu, X, Zhao, J, Wang, H, Peng, Y, Li, P, Li, Y. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.051 Å) | | Cite: | Crystal structure of Peroxiredoxin-1 from Schistosoma japonicum
To Be Published
|
|
5DH3
 
 | | Crystal structure of MST2 in complex with XMU-MP-1 | | Descriptor: | 4-[(5,10-dimethyl-6-oxo-6,10-dihydro-5H-pyrimido[5,4-b]thieno[3,2-e][1,4]diazepin-2-yl)amino]benzenesulfonamide, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2015-08-29 | | Release date: | 2016-08-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration
Sci Transl Med, 8, 2016
|
|
6TQL
 
 | | Cryo-EM of elastase-treated human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
6TQK
 
 | | Cryo-EM of native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
4RZE
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, L437W,D594E mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Fengwei, L, Xuyang, T, Anzhong, L, Li, L, Yuan, L, Hangzi, C, Qiao, W, Tianwei, L. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
7PFP
 
 | | Full-length cryo-EM structure of the native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-08-11 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6U4Y
 
 | | Crystal Structure of an EZH2-EED Complex in an Oligomeric State | | Descriptor: | Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Jiao, L, Liu, X. | | Deposit date: | 2019-08-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A partially disordered region connects gene repression and activation functions of EZH2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6V2F
 
 | | Crystal structure of the HIV capsid hexamer bound to the small molecule long-acting inhibitor, GS-6207 | | Descriptor: | HIV-1 capsid, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Appleby, T.C, Link, J.O, Yant, S.R, Villasenor, A.G, Somoza, J.R, Hu, E.Y, Schroeder, S.D, Cihlar, T. | | Deposit date: | 2019-11-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Clinical targeting of HIV capsid protein with a long-acting small molecule.
Nature, 584, 2020
|
|
