1TWS
 
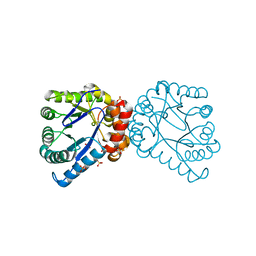 | | Dihydropteroate Synthetase From Bacillus anthracis | | Descriptor: | DHPS, Dihydropteroate synthase, SULFATE ION | | Authors: | Babaoglu, K, Qi, J, Lee, R.E, White, S.W. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of 7,8-Dihydropteroate Synthase from Bacillus anthracis; Mechanism and Novel Inhibitor Design.
STRUCTURE, 12, 2004
|
|
1TX2
 
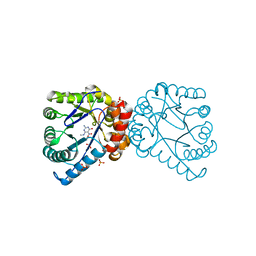 | | Dihydropteroate Synthetase, With Bound Inhibitor MANIC, From Bacillus anthracis | | Descriptor: | 6-METHYLAMINO-5-NITROISOCYTOSINE, DHPS, Dihydropteroate synthase, ... | | Authors: | Babaoglu, K, Qi, J, Lee, R.E, White, S.W. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of 7,8-Dihydropteroate Synthase from Bacillus anthracis; Mechanism and Novel Inhibitor Design.
STRUCTURE, 12, 2004
|
|
1TWW
 
 | | Dihydropteroate Synthetase, With Bound Substrate Analogue PtPP, From Bacillus anthracis | | Descriptor: | 6-HYDROXYMETHYLPTERIN-DIPHOSPHATE, DHPS, Dihydropteroate synthase, ... | | Authors: | Babaoglu, K, Qi, J, Lee, R.E, White, S.W. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of 7,8-Dihydropteroate Synthase from Bacillus anthracis; Mechanism and Novel Inhibitor Design.
STRUCTURE, 12, 2004
|
|
7K77
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ001008025 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-09-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ001008025
To Be Published
|
|
3TYE
 
 | | Dihydropteroate Synthase in complex with DHP-STZ | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, 4-amino-N-(1,3-thiazol-2-yl)benzenesulfonamide, 4-{[(2-amino-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)methyl]amino}-N-(1,3-thiazol-2-yl)benzenesulfonamide, ... | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2011-09-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalysis and sulfa drug resistance in dihydropteroate synthase.
Science, 335, 2012
|
|
3TS3
 
 | |
3TYC
 
 | | Dihydropteroate Synthase in complex with DHP+ | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, Dihydropteroate synthase, SULFATE ION | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2011-09-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Catalysis and sulfa drug resistance in dihydropteroate synthase.
Science, 335, 2012
|
|
3TYD
 
 | | Dihydropteroate Synthase in complex with PPi and DHP+ | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, Dihydropteroate synthase, PYROPHOSPHATE 2-, ... | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2011-09-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalysis and sulfa drug resistance in dihydropteroate synthase.
Science, 335, 2012
|
|
3S12
 
 | | Crystal structure of H5N1 influenza virus hemagglutinin, strain YU562 crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | DuBois, R.M, Zaraket, H, Reddivari, M, Heath, R.J, White, S.W, Russell, C.J. | | Deposit date: | 2011-05-14 | | Release date: | 2011-12-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Acid stability of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity.
Plos Pathog., 7, 2011
|
|
3SQ7
 
 | |
3SQ5
 
 | |
3SQ3
 
 | |
3UPU
 
 | | Crystal structure of the T4 Phage SF1B Helicase Dda | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*T)-3', ATP-dependent DNA helicase dda | | Authors: | He, X, Yun, M.K, Pemble IV, C.W, Kreuzer, K.N, Raney, K.D, White, S.W. | | Deposit date: | 2011-11-18 | | Release date: | 2012-06-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | The T4 Phage SF1B Helicase Dda Is Structurally Optimized to Perform DNA Strand Separation.
Structure, 20, 2012
|
|
3TYB
 
 | | Dihydropteroate Synthase in complex with pHBA and DHP+ | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, Dihydropteroate synthase, P-HYDROXYBENZOIC ACID, ... | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2011-09-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis and sulfa drug resistance in dihydropteroate synthase.
Science, 335, 2012
|
|
6V6X
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000988632 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, N-[2-(6-amino-9H-purin-9-yl)ethyl]-5-hydroxy-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
3SQ8
 
 | |
3S11
 
 | | Crystal structure of H5N1 influenza virus hemagglutinin, strain 437-10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | DuBois, R.M, Zaraket, H, Reddivari, M, Heath, R.J, White, S.W, Russell, C.J. | | Deposit date: | 2011-05-14 | | Release date: | 2011-12-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Acid stability of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity.
Plos Pathog., 7, 2011
|
|
6VG9
 
 | | The crystal structure of the 2009 H1N1/California PA endonuclease I38T mutant in complex with SJ000986248 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-07 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6V9E
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease wild type in complex with SJ000988632 | | Descriptor: | 1,1',1'',1''',1''''-[(3R,5S,7S,9R)-decane-1,3,5,7,9-pentayl]penta(pyrrolidin-2-one), MANGANESE (II) ION, N-[2-(6-amino-9H-purin-9-yl)ethyl]-5-hydroxy-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-13 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VBR
 
 | | The crystal structure of the 2009 H1N1/California PA endonuclease wild type in complex with SJ000986248 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-19 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
3V5O
 
 | | Structural and Mechanistic Studies of Catalysis and Sulfa Drug Resistance in Dihydropteroate Synthase | | Descriptor: | Dihydropteroate synthase, SULFATE ION | | Authors: | Yun, M, Wu, Y, Li, Z, Zhao, Y, Waddell, M.B, Ferreira, A.M, Lee, R.E, Bashford, D, White, S.W. | | Deposit date: | 2011-12-16 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalysis and sulfa drug resistance in dihydropteroate synthase.
Science, 335, 2012
|
|
1WHI
 
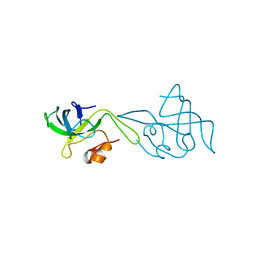 | | RIBOSOMAL PROTEIN L14 | | Descriptor: | RIBOSOMAL PROTEIN L14 | | Authors: | Davies, C, White, S.W, Ramakrishnan, V. | | Deposit date: | 1996-01-10 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of ribosomal protein L14 reveals an important organizational component of the translational apparatus.
Structure, 4, 1996
|
|
1PKP
 
 | |
1I1S
 
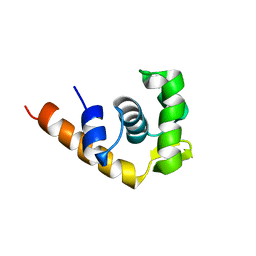 | | SOLUTION STRUCTURE OF THE TRANSCRIPTIONAL ACTIVATION DOMAIN OF THE BACTERIOPHAGE T4 PROTEIN MOTA | | Descriptor: | MOTA | | Authors: | Li, N, Zhang, W, White, S.W, Kriwacki, R.W. | | Deposit date: | 2001-02-02 | | Release date: | 2001-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the transcriptional activation domain of the bacteriophage T4 protein, MotA.
Biochemistry, 40, 2001
|
|
1I01
 
 | |
