6HUZ
 
 | | HmdII from Desulfurobacterium thermolithotrophum reconstituted with Fe-guanylylpyridinol (FeGP) cofactor and co-crystallized with methenyl-tetrahydrofolate form B | | Descriptor: | 1,2-ETHANEDIOL, 5,10-Methenyltetrahydrofolate, Coenzyme F420-dependent N(5),N(10)-methenyltetrahydromethanopterin reductase-related protein, ... | | Authors: | Watanabe, T, Wagner, T, Huang, G, Kahnt, J, Ataka, K, Ermler, U, Shima, S. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Bacterial [Fe]-Hydrogenase Paralog HmdII Uses Tetrahydrofolate Derivatives as Substrates.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6HUY
 
 | | HmdII from Desulfurobacterium thermolithotrophum reconstitued with Fe-guanylylpyridinol (FeGP) cofactor and co-crystallized with methenyl-tetrahydrofolate form A | | Descriptor: | 5,10-Methenyltetrahydrofolate, Coenzyme F420-dependent N(5),N(10)-methenyltetrahydromethanopterin reductase-related protein, DIMETHYL SULFOXIDE, ... | | Authors: | Watanabe, T, Wagner, T, Huang, G, Kahnt, J, Ataka, K, Ermler, U, Shima, S. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Bacterial [Fe]-Hydrogenase Paralog HmdII Uses Tetrahydrofolate Derivatives as Substrates.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6HUX
 
 | | HmdII from Methanocaldococcus jannaschii reconstitued with Fe-guanylylpyridinol (FeGP) cofactor and co-crystallized with methenyl-tetrahydromethanopterin at 2.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[(6S,6aR,7R)-3-amino-6,7-dimethyl-1-oxo-1,2,5,6,6a,7-hexahydro-8H-imidazo[1,5-f]pteridin-10-ium-8-yl]phenyl}-1-deoxy-5-O-{5-O-[(S)-{[(1S)-1,3-dicarboxypropyl]oxy}(hydroxy)phosphoryl]-alpha-D-ribofuranosyl}-D-ribitol, ACETATE ION, ... | | Authors: | Watanabe, T, Wagner, T, Huang, G, Kahnt, J, Ataka, K, Ermler, U, Shima, S. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Bacterial [Fe]-Hydrogenase Paralog HmdII Uses Tetrahydrofolate Derivatives as Substrates.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
7BKD
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (heterodislfide reductase core and mobile arm in conformational state 1, composite structure) | | Descriptor: | CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, CoB--CoM heterodisulfide reductase subunit C, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKE
 
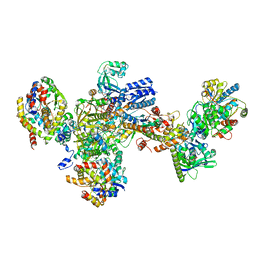 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (heterodisulfide reductase core and mobile arm in conformational state 2, composite structure) | | Descriptor: | CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, CoB--CoM heterodisulfide reductase subunit C, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKB
 
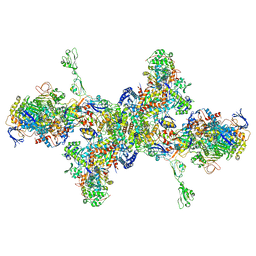 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (hexameric, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKC
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (dimeric, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
1F2E
 
 | | STRUCTURE OF SPHINGOMONAD, GLUTATHIONE S-TRANSFERASE COMPLEXED WITH GLUTATHIONE | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Nishio, T, Watanabe, T, Patel, A, Wang, Y, Lau, P.C.K, Grochulski, P, Li, Y, Cygler, M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-06-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Properties of a Sphingomonad and Marine Bacterium Beta-Class Glutathione S-Transferases and Crystal Structure of the Former Complex with Glutathione
To be published
|
|
3IWR
 
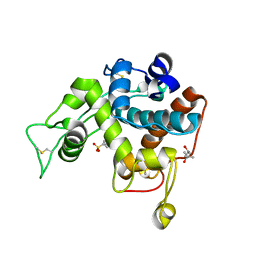 | | Crystal structure of class I chitinase from Oryza sativa L. japonica | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chitinase | | Authors: | Kezuka, Y, Watanabe, T, Nonaka, T. | | Deposit date: | 2009-09-03 | | Release date: | 2010-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of full-length class I chitinase from rice revealed by X-ray crystallography and small-angle X-ray scattering.
Proteins, 78, 2010
|
|
1ITX
 
 | | Catalytic Domain of Chitinase A1 from Bacillus circulans WL-12 | | Descriptor: | GLYCEROL, Glycosyl Hydrolase | | Authors: | Iwahori, F, Matsumoto, T, Watanabe, T, Nonaka, T. | | Deposit date: | 2002-02-13 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Three-dimensional structure of the catalytic domain of chitinase A1 from Bacillus circulans WL-12 at a very high resolution
PROC.JPN.ACAD.,SER.B, 75, 1999
|
|
1WVU
 
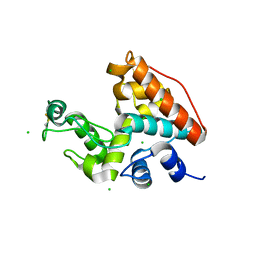 | |
1WVV
 
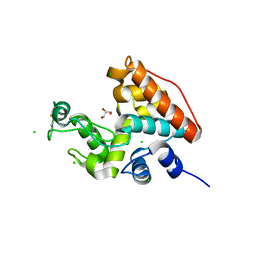 | |
4X68
 
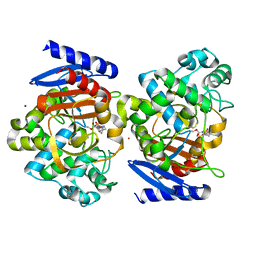 | | Crystal Structure of OP0595 complexed with AmpC | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, NICKEL (II) ION | | Authors: | Yamada, M, Watanabe, T. | | Deposit date: | 2014-12-07 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | OP0595, a new diazabicyclooctane: mode of action as a serine beta-lactamase inhibitor, antibiotic and beta-lactam 'enhancer'
J.Antimicrob.Chemother., 70, 2015
|
|
4X69
 
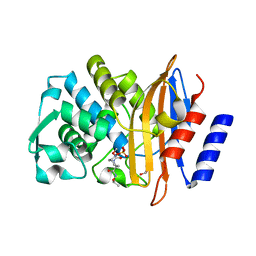 | | Crystal structure of OP0595 complexed with CTX-M-44 | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase Toho-1 | | Authors: | Yamada, M, Watanabe, T. | | Deposit date: | 2014-12-07 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | OP0595, a new diazabicyclooctane: mode of action as a serine beta-lactamase inhibitor, antibiotic and beta-lactam 'enhancer'
J.Antimicrob.Chemother., 70, 2015
|
|
1ED7
 
 | | SOLUTION STRUCTURE OF THE CHITIN-BINDING DOMAIN OF BACILLUS CIRCULANS WL-12 CHITINASE A1 | | Descriptor: | CHITINASE A1 | | Authors: | Ikegami, T, Okada, T, Hashimoto, M, Seino, S, Watanabe, T, Shirakawa, M. | | Deposit date: | 2000-01-27 | | Release date: | 2000-05-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1.
J.Biol.Chem., 275, 2000
|
|
5B2H
 
 | | Crystal structure of HA33 from Clostridium botulinum serotype C strain Yoichi | | Descriptor: | HA-33, TRIETHYLENE GLYCOL | | Authors: | Akiyama, T, Hayashi, S, Matsumoto, T, Hasegawa, K, Yamano, A, Suzuki, T, Niwa, K, Watanabe, T, Sagane, Y, Yajima, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational divergence in the HA-33/HA-17 trimer of serotype C and D botulinum toxin complex
Biochem.Biophys.Res.Commun., 476, 2016
|
|
2DBT
 
 | |
8GZ6
 
 | | Crystal structure of neutralizing VHH P17 in complex with SARS-CoV-2 Alpha variant spike receptor-binding domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nanobody P17 | | Authors: | Yamaguchi, K, Anzai, I, Maeda, R, Moriguchi, M, Watanabe, T, Imura, A, Takaori-Kondo, A, Inoue, T. | | Deposit date: | 2022-09-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the rational design of a nanobody that binds with high affinity to the SARS-CoV-2 spike variant.
J.Biochem., 173, 2023
|
|
8GZ5
 
 | | Crystal structure of neutralizing VHH P17 in complex with SARS-CoV-2 Alpha variant spike receptor-binding domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P17, ... | | Authors: | Yamaguchi, K, Anzai, I, Maeda, R, Moriguchi, M, Watanabe, T, Imura, A, Takaori-Kondo, A, Inoue, T. | | Deposit date: | 2022-09-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the rational design of a nanobody that binds with high affinity to the SARS-CoV-2 spike variant.
J.Biochem., 173, 2023
|
|
2D49
 
 | | Solution structure of the Chitin-Binding Domain of Streptomyces griseus Chitinase C | | Descriptor: | chitinase C | | Authors: | Akagi, K, Watanabe, J, Hara, M, Kezuka, Y, Chikaishi, E, Yamaguchi, T, Akutsu, H, Nonaka, T, Watanabe, T, Ikegami, T. | | Deposit date: | 2005-10-11 | | Release date: | 2006-10-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Identification of the substrate interaction region of the chitin-binding domain of Streptomyces griseus chitinase C
J.Biochem.(Tokyo), 139, 2006
|
|
2DKV
 
 | | Crystal structure of class I chitinase from Oryza sativa L. japonica | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, chitinase | | Authors: | Kezuka, Y, Nishizawa, Y, Watanabe, T, Nonaka, T. | | Deposit date: | 2006-04-14 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of full-length class I chitinase from rice revealed by X-ray crystallography and small-angle X-ray scattering.
Proteins, 78, 2010
|
|
7CC4
 
 | |
1K85
 
 | | Solution structure of the fibronectin type III domain from Bacillus circulans WL-12 Chitinase A1. | | Descriptor: | CHITINASE A1 | | Authors: | Jee, J.G, Ikegami, T, Hashimoto, M, Kawabata, T, Ikeguchi, M, Watanabe, T, Shirakawa, M. | | Deposit date: | 2001-10-23 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Fibronectin Type III Domain
from Bacillus circulans WL-12 Chitinase A1
J.Biol.Chem., 277, 2002
|
|
2RTT
 
 | | Solution structure of the chitin-binding domain of Chi18aC from Streptomyces coelicolor | | Descriptor: | ChiC | | Authors: | Okumura, A, Uemura, M, Yamada, N, Chikaishi, E, Takai, T, Yoshio, S, Akagi, K, Morita, J, Lee, Y, Yokogawa, D, Suzuki, K, Watanabe, T, Ikegami, T. | | Deposit date: | 2013-08-26 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Chitin-binding domain of chitinase Chi18aC from Streptomyces coelicolor
To be Published
|
|
6NBI
 
 | | Cryo-EM structure of parathyroid hormone receptor type 1 in complex with a long-acting parathyroid hormone analog and G protein | | Descriptor: | CHOLESTEROL, Gs protein alpha subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, L.-H, Ma, S, Sutkeviciute, I, Shen, D.-D, Zhou, X.E, de Waal, P.P, Li, C.-Y, Kang, Y, Clark, L.J, Jean-Alphonse, F.G, White, A.D, Xiao, K, Yang, D, Jiang, Y, Watanabe, T, Gardella, T.J, Melcher, K, Wang, M.-W, Vilardaga, J.-P, Xu, H.E, Zhang, Y. | | Deposit date: | 2018-12-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and dynamics of the active human parathyroid hormone receptor-1.
Science, 364, 2019
|
|
