4F3O
 
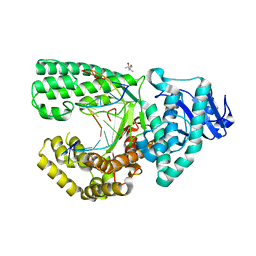 | | DNA Polymerase I Large Fragment Complex 5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*C*AP*TP*GP*AP*GP*AP*GP*TP*CP*AP*GP*G)-3'), ... | | Authors: | Wang, W, Beese, L.S. | | Deposit date: | 2012-05-09 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structures of a High-fidelity DNA Polymerase
to be published
|
|
4F8R
 
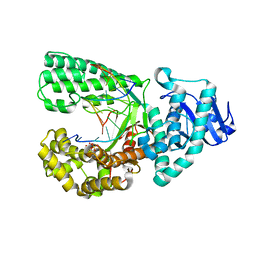 | | Bacillus DNA Polymerase I Large Fragment complex 7 | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*CP*GP*AP*GP*TP*CP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DDG))-3'), DNA polymerase, ... | | Authors: | Wang, W, Beese, L.S. | | Deposit date: | 2012-05-17 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of a High-fidelity DNA Polymerase
to be published
|
|
4EZ6
 
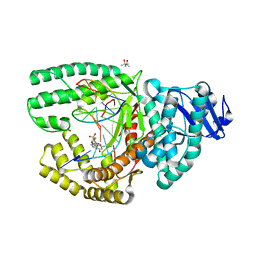 | | Bacillus DNA Polymerase I Large Fragment Complex 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*TP*GP*CP*GP*AP*GP*TP*CP*AP*GP*G)-3'), ... | | Authors: | Wang, W, Beese, L.S. | | Deposit date: | 2012-05-02 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of a High-fidelity DNA Polymerase
to be published
|
|
4F4K
 
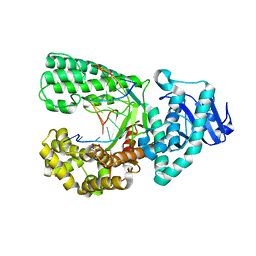 | | DNA Polymerase I Large Fragment Complex 6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*TP*TP*CP*GP*AP*GP*TP*CP*AP*GP*G)-3'), ... | | Authors: | Wang, W, Beese, L.S. | | Deposit date: | 2012-05-10 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of a High-fidelity DNA Polymerase
to be published
|
|
4F2R
 
 | | DNA Polymerase I Large Fragment complex 3 | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*TP*GP*GP*GP*AP*GP*TP*CP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DOC))-3'), ... | | Authors: | Wang, W, Beese, L.S. | | Deposit date: | 2012-05-08 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structures of a High-fidelity DNA Polymerase
to be published
|
|
3PX0
 
 | |
3PV8
 
 | |
3PX6
 
 | |
3PS5
 
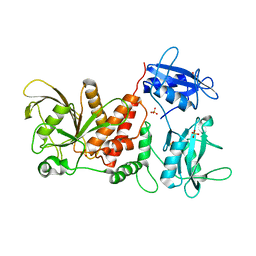 | | Crystal structure of the full-length Human Protein Tyrosine Phosphatase SHP-1 | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 6 | | Authors: | Wang, W, Liu, L, Song, X, Mo, Y, Komma, C, Bellamy, H.D, Zhao, Z.J, Zhou, G.W. | | Deposit date: | 2010-11-30 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of human protein tyrosine phosphatase SHP-1 in the open conformation.
J.Cell.Biochem., 112, 2011
|
|
3PX4
 
 | |
7TO1
 
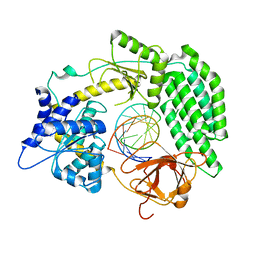 | | Cryo-EM structure of RIG-I bound to the end of p3SLR30 (+ATP) | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p3SLR30 | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNY
 
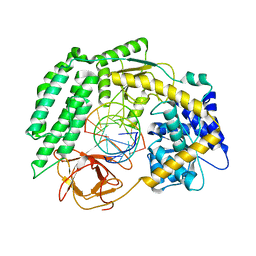 | | Cryo-EM structure of RIG-I in complex with p2dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p2dsRNA | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TO2
 
 | |
7TO0
 
 | | Cryo-EM structure of RIG-I in complex with OHdsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, OHdsRNA, ZINC ION | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNZ
 
 | | Cryo-EM structure of RIG-I in complex with p1dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p1dsRNA | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNX
 
 | | Cryo-EM structure of RIG-I in complex with p3dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p3dsRNAa, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
8G7T
 
 | | Cryo-EM structure of RNP end | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-inter)
To Be Published
|
|
8G7U
 
 | | Cryo-EM structure of RNP end 2 | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-inter)
To Be Published
|
|
8G7V
 
 | | Cryo-EM structure of RNP inter | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-inter)
To Be Published
|
|
5XGQ
 
 | |
5XET
 
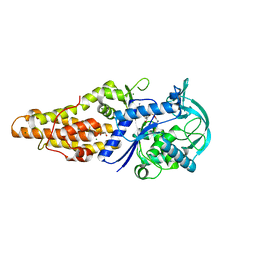 | | Crystal structure of Mycobacterium tuberculosis methionyl-tRNA synthetase bound by methionyl-adenylate (Met-AMP) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methionine--tRNA ligase, ... | | Authors: | Wang, W, Qin, B, Wojdyla, J.A, Wang, M, Gao, X, Cui, S. | | Deposit date: | 2017-04-06 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural characterization of free-state and product-stateMycobacterium tuberculosismethionyl-tRNA synthetase reveals an induced-fit ligand-recognition mechanism.
IUCrJ, 5, 2018
|
|
5Z1Q
 
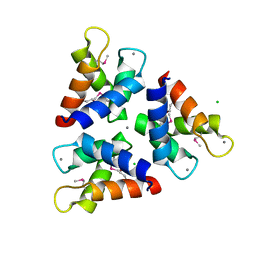 | | Crystal structures of the trimeric N-terminal domain of Ciliate Euplotes octocarinatus Centrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Centrin protein | | Authors: | Wang, W, Zhao, Y, Yang, B, Wang, H. | | Deposit date: | 2017-12-27 | | Release date: | 2018-04-25 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the trimeric N-terminal domain of ciliate Euplotes octocarinatus centrin binding with calcium ions
Protein Sci., 27, 2018
|
|
6BLP
 
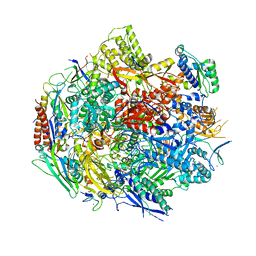 | | Pol II elongation complex with an abasic lesion at i+1 position, soaking AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA (5'-D(P*AP*(3DR)P*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, W, Wang, D. | | Deposit date: | 2017-11-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BM2
 
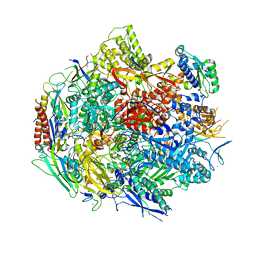 | | Pol II elongation complex with an abasic lesion at i-1 position | | Descriptor: | DNA (5'-D(P*CP*AP*(3DR)P*CP*TP*CP*TP*TP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Wang, W, Wang, D. | | Deposit date: | 2017-11-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.403 Å) | | Cite: | Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BM4
 
 | | Pol II elongation complex with an abasic lesion at i-1 position,soaking UMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, DNA (5'-D(P*CP*AP*(3DR)P*CP*TP*CP*TP*TP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, W, Wang, D. | | Deposit date: | 2017-11-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
