3GJB
 
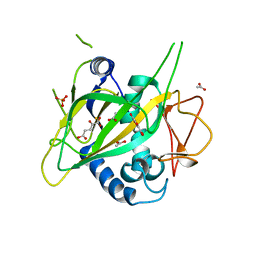 | | CytC3 with Fe(II) and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, CytC3, ... | | Authors: | Wong, C, Drennan, C.L. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of an open active site conformation of nonheme iron halogenase CytC3
J.Am.Chem.Soc., 131, 2009
|
|
3GJA
 
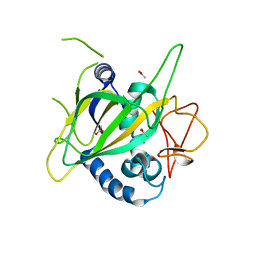 | | CytC3 | | Descriptor: | ACETATE ION, CytC3 | | Authors: | Wong, C, Drennan, C.L. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of an open active site conformation of nonheme iron halogenase CytC3
J.Am.Chem.Soc., 131, 2009
|
|
2DLN
 
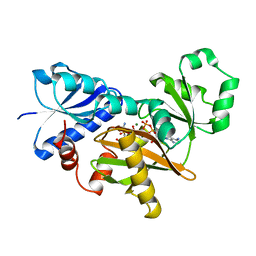 | | VANCOMYCIN RESISTANCE: STRUCTURE OF D-ALANINE:D-ALANINE LIGASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 1(S)-AMINOETHYL-(2-CARBOXYPROPYL)PHOSPHORYL-PHOSPHINIC ACID, ADENOSINE-5'-DIPHOSPHATE, D-ALANINE--D-ALANINE LIGASE, ... | | Authors: | Knox, J.R, Moews, P.C, Fan, C. | | Deposit date: | 1994-07-18 | | Release date: | 1995-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Vancomycin resistance: structure of D-alanine:D-alanine ligase at 2.3 A resolution.
Science, 266, 1994
|
|
2E4G
 
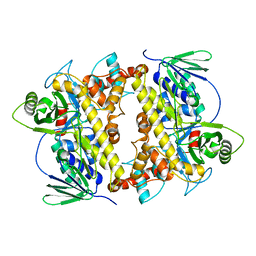 | | RebH with bound L-Trp | | Descriptor: | TRYPTOPHAN, Tryptophan halogenase | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2006-12-07 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases
Biochemistry, 46, 2007
|
|
1OFN
 
 | | Purification, crystallisation and preliminary structural studies of dTDP-4-keto-6-deoxy-glucose-5-epimerase (EvaD) from Amycolatopsis orientalis; the fourth enzyme in the dTDP-L-epivancosamine biosynthetic pathway. | | Descriptor: | GLYCEROL, PCZA361.16 | | Authors: | Merkel, A.B, Naismith, J.H. | | Deposit date: | 2003-04-17 | | Release date: | 2004-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Purification, Crystallization and Preliminary Structural Studies of Dtdp-4-Keto-6-Deoxy-Glucose-5-Epimerase (Evad) from Amycolatopsis Orientalis, the Fourth Enzyme in the Dtdp-L-Epivancosamine Biosynthetic Pathway.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3BMZ
 
 | | Violacein biosynthetic enzyme VioE | | Descriptor: | Putative uncharacterized protein, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-12-13 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The Violacein Biosynthetic Enzyme VioE Shares a Fold with Lipoprotein Transporter Proteins
J.Biol.Chem., 283, 2008
|
|
3EPT
 
 | |
1BD0
 
 | | ALANINE RACEMASE COMPLEXED WITH ALANINE PHOSPHONATE | | Descriptor: | ALANINE RACEMASE, {1-[(3-HYDROXY-METHYL-5-PHOSPHONOOXY-METHYL-PYRIDIN-4-YLMETHYL)-AMINO]-ETHYL}-PHOSPHONIC ACID | | Authors: | Stamper, G.F, Morollo, A.A, Ringe, D. | | Deposit date: | 1998-05-12 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reaction of alanine racemase with 1-aminoethylphosphonic acid forms a stable external aldimine.
Biochemistry, 37, 1998
|
|
1CLH
 
 | |
4Z2Y
 
 | | Crystal structure of methyltransferase CalO6 | | Descriptor: | CalO6, MERCURY (II) ION | | Authors: | Hou, C, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of O-methyltransferase CalO6 from the calicheamicin biosynthetic pathway: a case of challenging structure determination at low resolution.
Bmc Struct.Biol., 15, 2015
|
|
2OAL
 
 | | RebH with bound FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Tryptophan halogenase | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2006-12-16 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases
Biochemistry, 46, 2007
|
|
2OAM
 
 | |
2R0G
 
 | | Chromopyrrolic acid-soaked RebC with bound 7-carboxy-K252c | | Descriptor: | 7-carboxy-5-hydroxy-12,13-dihydro-6H-indolo[2,3-a]pyrrolo[3,4-c]carbazole, FLAVIN-ADENINE DINUCLEOTIDE, RebC | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-08-19 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R0P
 
 | | K252c-soaked RebC | | Descriptor: | 6,7,12,13-tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazol-5-one, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-08-20 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R0C
 
 | |
2RMA
 
 | |
2RMB
 
 | |
2FCT
 
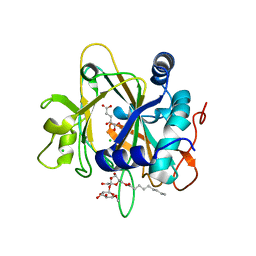 | | SyrB2 with Fe(II), chloride, and alpha-ketoglutarate | | Descriptor: | ((2R,3S,4S,5S)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)-5-((2R,3S,4S,5S,6R)-3,4,5-TRIHYDROXY-6-METHOXY-TETRAHYDRO-2H-PYRAN-2-YLOXY)-TETRAHYDROFURAN-2-YL)METHYL NONANOATE, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis.
Nature, 440, 2006
|
|
2FCV
 
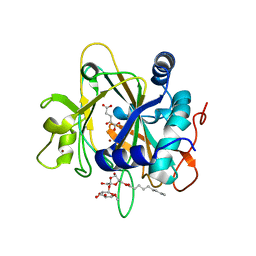 | | SyrB2 with Fe(II), bromide, and alpha-ketoglutarate | | Descriptor: | ((2R,3S,4S,5S)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)-5-((2R,3S,4S,5S,6R)-3,4,5-TRIHYDROXY-6-METHOXY-TETRAHYDRO-2H-PYRAN-2-YLOXY)-TETRAHYDROFURAN-2-YL)METHYL NONANOATE, 2-OXOGLUTARIC ACID, BROMIDE ION, ... | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis.
Nature, 440, 2006
|
|
2FCU
 
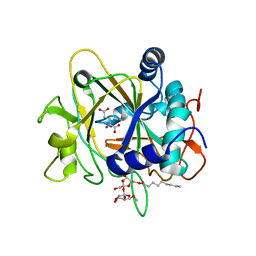 | | SyrB2 with alpha-ketoglutarate | | Descriptor: | ((2R,3S,4S,5S)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)-5-((2R,3S,4S,5S,6R)-3,4,5-TRIHYDROXY-6-METHOXY-TETRAHYDRO-2H-PYRAN-2-YLOXY)-TETRAHYDROFURAN-2-YL)METHYL NONANOATE, 2-OXOGLUTARIC ACID, syringomycin biosynthesis enzyme 2 | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis.
Nature, 440, 2006
|
|
2CPL
 
 | |
1NDA
 
 | | THE STRUCTURE OF TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE IN THE OXIDIZED AND NADPH REDUCED STATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, TRYPANOTHIONE OXIDOREDUCTASE | | Authors: | Lantwin, C.B, Kabsch, W, Pai, E.F, Schlichting, I, Krauth-Siegel, R.L. | | Deposit date: | 1993-07-02 | | Release date: | 1994-09-30 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of Trypanosoma cruzi trypanothione reductase in the oxidized and NADPH reduced state.
Proteins, 18, 1994
|
|
1QFL
 
 | |
1SFT
 
 | | ALANINE RACEMASE | | Descriptor: | ACETATE ION, ALANINE RACEMASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shaw, J.P, Petsko, G.A, Ringe, D. | | Deposit date: | 1996-09-20 | | Release date: | 1997-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-A resolution.
Biochemistry, 36, 1997
|
|
1UAE
 
 | | STRUCTURE OF UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE | | Descriptor: | UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, [(1R)-1-hydroxypropyl]phosphonic acid | | Authors: | Skarzynski, T. | | Deposit date: | 1996-09-30 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin.
Structure, 4, 1996
|
|
