6JD9
 
 | | Proteus mirabilis lipase mutant - I118V/E130G | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase, CALCIUM ION | | Authors: | Heater, B.S, Chan, W.S, Chan, M.K. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Directed evolution of a genetically encoded immobilized lipase for the efficient production of biodiesel from waste cooking oil.
Biotechnol Biofuels, 12, 2019
|
|
6IWY
 
 | |
6KNT
 
 | | Crystal structure of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis (space group P4332) | | Descriptor: | Putative metal-dependent hydrolase, ZINC ION | | Authors: | Na, H.W, Namgung, B, Song, W.S, Yoon, S.I. | | Deposit date: | 2019-08-07 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
3S84
 
 | | Dimeric apoA-IV | | Descriptor: | Apolipoprotein A-IV, SULFATE ION | | Authors: | Deng, X, Davidson, W.S, Thompson, T.B. | | Deposit date: | 2011-05-27 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of Dimeric Apolipoprotein A-IV and Its Mechanism of Self-Association.
Structure, 20, 2012
|
|
1HKG
 
 | |
3V3X
 
 | | Nitroxide Spin Labels in Protein GB1: N8/K28 Double Mutant | | Descriptor: | ACETATE ION, GLYCEROL, Immunoglobulin G-binding protein G, ... | | Authors: | Cunningham, T.F, McGoff, M.S, Sengupta, I, Jaroniec, C.P, Horne, W.S, Saxena, S.K. | | Deposit date: | 2011-12-14 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a protein spin-label in a solvent-exposed beta-sheet and comparison with DEER spectroscopy.
Biochemistry, 51, 2012
|
|
7RAP
 
 | |
7TIO
 
 | |
7TIP
 
 | |
7TIS
 
 | |
7TIQ
 
 | |
7TIR
 
 | |
7TV6
 
 | | Heterogeneous-backbone proteomimetic analogue of the disulfide-rich venom peptide lasiocepsin: native loop | | Descriptor: | Lasiocepsin heterogeneous-backbone proteomimetic analogue | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
3ZXJ
 
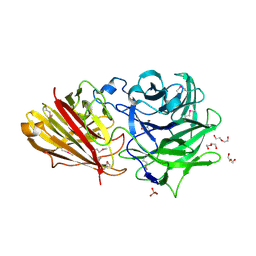 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HIAXHD3, ... | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZXK
 
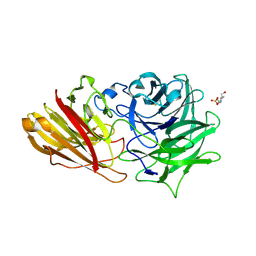 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIAXHD3, alpha-L-arabinofuranose-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7TV8
 
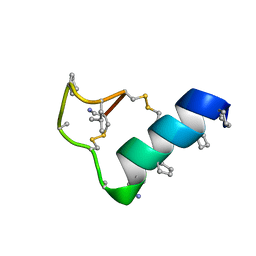 | | Heterogeneous-backbone proteomimetic analogue of the disulfide-rich venom peptide lasiocepsin: D-Ala modified loop | | Descriptor: | Lasiocepsin heterogeneous-backbone proteomimetic analogue | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
7TV5
 
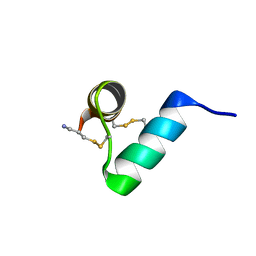 | | Disulfide-rich venom peptide lasiocepsin: P20A mutant | | Descriptor: | Lasiocepsin | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
7TV7
 
 | | Heterogeneous-backbone proteomimetic analogue of the disulfide-rich venom peptide lasiocepsin: beta-3-Lys modified loop | | Descriptor: | Lasiocepsin heterogeneous-backbone proteomimetic analogue | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
3ZXL
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HIAXHD3 | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4A1U
 
 | | Crystal structure of alpha-beta-foldamer 2c in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-FOLDAMER 2C, BCL-2-LIKE PROTEIN 1, CHLORIDE ION, ... | | Authors: | Boersma, M.D, Haase, H.S, Kaufman, K.J, Horne, W.S, Lee, E.F, Clarke, O.B, Smith, B.J, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-09-20 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Evaluation of Diverse Alpha/Beta-Backbone Patterns for Functional Alpha-Helix Mimicry: Analogues of the Bim Bh3 Domain.
J.Am.Chem.Soc., 134, 2012
|
|
4A1W
 
 | | Crystal structure of alpha-beta foldamer 4c in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-FOLDAMER 2C, BCL-2-LIKE PROTEIN 1 | | Authors: | Boersma, M.D, Haase, H.S, Kaufman, K.J, Horne, W.S, Lee, E.F, Clarke, O.B, Smith, B.J, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-09-20 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Evaluation of Diverse Alpha/Beta-Backbone Patterns for Functional Alpha-Helix Mimicry: Analogues of the Bim Bh3 Domain.
J.Am.Chem.Soc., 134, 2012
|
|
1CMA
 
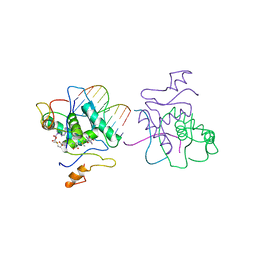 | | MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE | | Descriptor: | DNA (5'-D(*AP*GP*AP*CP*GP*TP*CP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*AP*CP*GP*TP*CP*T)-3'), PROTEIN (MET REPRESSOR), ... | | Authors: | Somers, W.S, Phillips, S.E.V. | | Deposit date: | 1992-08-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands.
Nature, 359, 1992
|
|
1DII
 
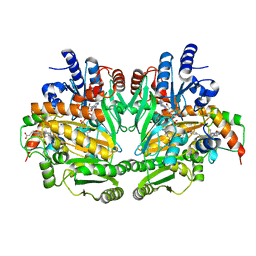 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE AT 2.5 A RESOLUTION | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | Authors: | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.N, McIntire, W.S. | | Deposit date: | 1999-11-29 | | Release date: | 1999-12-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
1DIQ
 
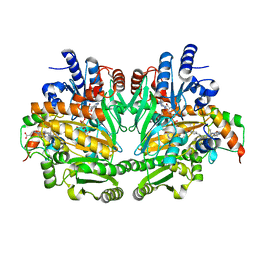 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE WITH SUBSTRATE BOUND | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | Authors: | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.S, McIntire, W.S. | | Deposit date: | 1999-11-29 | | Release date: | 1999-12-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
7CKC
 
 | | Simplified Alpha-Carboxysome, T=4 | | Descriptor: | Major carboxysome shell protein 1A, Unidentified carboxysome polypeptide | | Authors: | Tan, Y.Q, Ali, S, Xue, B, Robinson, R.C, Narita, A, Yew, W.S. | | Deposit date: | 2020-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
