7SPP
 
 | |
7SPO
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with VNAR 3B4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2021-11-02 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Mechanisms of SARS-CoV-2 neutralization by shark variable new antigen receptors elucidated through X-ray crystallography.
Nat Commun, 12, 2021
|
|
7SMU
 
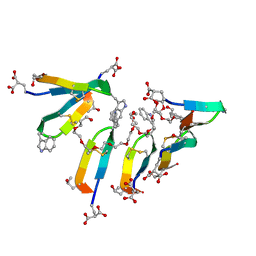 | | Crystal Structure of Consomatin-Ro1 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45-pentadecaoxaoctatetracontane-1,48-diol, Consomatin-Ro1 | | Authors: | Ramiro, I.B.L, Whitby, F.G, Hill, C.P, Safavi-Hemami, H, Concepcion, G.P, Olivera, B.M. | | Deposit date: | 2021-10-26 | | Release date: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Somatostatin venom analogs evolved by fish-hunting cone snails: From prey capture behavior to identifying drug leads.
Sci Adv, 8, 2022
|
|
7SZO
 
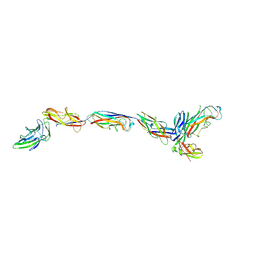 | | Structure of a bacterial fimbrial tip containing FocH | | Descriptor: | Chaperone protein FimC, FimF protein, FimG, ... | | Authors: | Stenkamp, R.E, Le Trong, I, Aprikian, P, Sokurenko, E.V. | | Deposit date: | 2021-11-29 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recombinant FimH Adhesin Demonstrates How the Allosteric Catch Bond Mechanism Can Support Fast and Strong Bacterial Attachment in the Absence of Shear.
J.Mol.Biol., 434, 2022
|
|
7SZ9
 
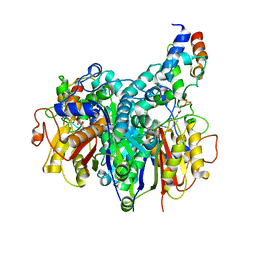 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C16:1-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N~3~-{(2R)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-N-(2-{[(9Z)-hexadec-9-enoyl]amino}ethyl)-beta-alaninamide, ... | | Authors: | Mindrebo, J.T, Chen, A, Noel, J.P, Burkart, M.D. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1AIZ
 
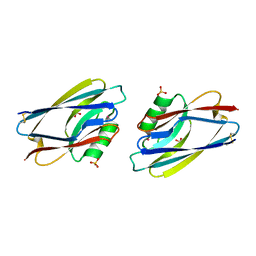 | | STRUCTURE OF APO-AZURIN FROM ALCALIGENES DENITRIFICANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, CADMIUM ION, SULFATE ION | | Authors: | Baker, E.N, Anderson, B.F, Blackwell, K.A. | | Deposit date: | 1993-11-11 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of apo-azurin from Alcaligenes denitrificans at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
2OXP
 
 | |
1AZC
 
 | | STRUCTURE OF APO-AZURIN FROM ALCALIGENES DENITRIFICANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, COPPER (II) ION, SULFATE ION | | Authors: | Baker, E.N, Shepard, W.E.B, Kingston, R.L. | | Deposit date: | 1992-12-16 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of apo-azurin from Alcaligenes denitrificans at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1AZB
 
 | |
2QSQ
 
 | | Crystal structure of the N-terminal domain of carcinoembryonic antigen (CEA) | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 5, GLYCEROL | | Authors: | Le Trong, I, Korotkova, N, Moseley, S.L, Stenkamp, R.E. | | Deposit date: | 2007-07-31 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding of Dr adhesins of Escherichia coli to carcinoembryonic antigen triggers receptor dissociation.
Mol.Microbiol., 67, 2008
|
|
2QST
 
 | |
1CM1
 
 | | MOTIONS OF CALMODULIN-SINGLE-CONFORMER REFINEMENT | | Descriptor: | CALCIUM ION, CALMODULIN, CALMODULIN-DEPENDENT PROTEIN KINASE II-ALPHA | | Authors: | Wall, M.E, Phillips Jr, G.N. | | Deposit date: | 1997-09-23 | | Release date: | 1998-03-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Motions of calmodulin characterized using both Bragg and diffuse X-ray scattering.
Structure, 5, 1997
|
|
1CM4
 
 | | Motions of calmodulin-four-conformer refinement | | Descriptor: | CALCIUM ION, CALMODULIN, CALMODULIN-DEPENDENT PROTEIN KINASE II-ALPHA | | Authors: | Wall, M.E, Phillips Jr, G.N. | | Deposit date: | 1997-09-23 | | Release date: | 1998-03-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Motions of calmodulin characterized using both Bragg and diffuse X-ray scattering.
Structure, 5, 1997
|
|
6ZOD
 
 | | Fusidic acid binding to the allosteric deep transmembrane domain binding pocket, TM7/TM8 groove, and TM1/TM2 groove of the fully induced AcrB T protomer | | Descriptor: | 1,2-ETHANEDIOL, DARPIN, DECANE, ... | | Authors: | Oswald, C, Tam, H.K, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
6PH4
 
 | | Full length LOV-PAS-HK construct from the LOV-HK sensory protein from Brucella abortus (light-adapted, construct 15-489) | | Descriptor: | Blue-light-activated histidine kinase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
4HTW
 
 | | SIVmac239 capsid N-terminal domain | | Descriptor: | Gag protein | | Authors: | Schmidt, A.G. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Gain-of-Sensitivity Mutations in a Trim5-Resistant Primary Isolate of Pathogenic SIV Identify Two Independent Conserved Determinants of Trim5alpha Specificity.
Plos Pathog., 9, 2013
|
|
4L4Y
 
 | |
4L4Z
 
 | | Crystal structures of the LsrR proteins complexed with phospho-AI-2 and its two different analogs reveal distinct mechanisms for ligand recognition | | Descriptor: | (2S)-2,3,3-trihydroxy-4-oxopentyl dihydrogen phosphate, Transcriptional regulator LsrR | | Authors: | Ryu, K.S, Ha, J.H, Eo, Y. | | Deposit date: | 2013-06-10 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the LsrR Proteins Complexed with Phospho-AI-2 and Two Signal-Interrupting Analogues Reveal Distinct Mechanisms for Ligand Recognition.
J.Am.Chem.Soc., 135, 2013
|
|
4L50
 
 | | Crystal structures of the LsrR proteins complexed with phospho-AI-2 and its two different analogs reveal distinct mechanisms for ligand recognition | | Descriptor: | (2S)-2,3,3-trihydroxy-6-methyl-4-oxoheptyl dihydrogen phosphate, Transcriptional regulator LsrR | | Authors: | Ryu, K.S, Ha, J.H, Eo, Y. | | Deposit date: | 2013-06-10 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the LsrR Proteins Complexed with Phospho-AI-2 and Two Signal-Interrupting Analogues Reveal Distinct Mechanisms for Ligand Recognition.
J.Am.Chem.Soc., 135, 2013
|
|
4L5I
 
 | |
4L6B
 
 | |
4L51
 
 | |
4L5J
 
 | |
4LAP
 
 | |
4LZ8
 
 | |
