6ENG
 
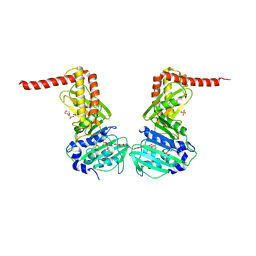 | | Crystal structure of the 43K ATPase domain of Escherichia coli gyrase B in complex with an aminocoumarin | | Descriptor: | CHLORIDE ION, Coumermycin A1, DNA gyrase subunit B, ... | | Authors: | Vanden Broeck, A, McEwen, A.G, Lamour, V. | | Deposit date: | 2017-10-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for DNA Gyrase Interaction with Coumermycin A1.
J.Med.Chem., 62, 2019
|
|
6ENH
 
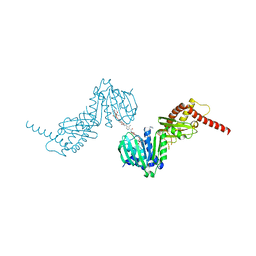 | |
6RKW
 
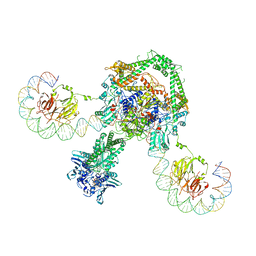 | | CryoEM structure of the complete E. coli DNA Gyrase complex bound to a 130 bp DNA duplex | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA (58-MER), DNA (62-MER), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
6RKS
 
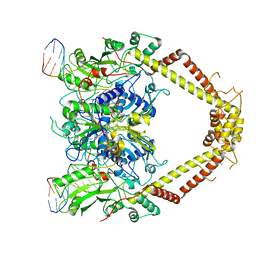 | | E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 without TOPRIM insertion | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA Strand 1, DNA Strand 2, ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
6RKV
 
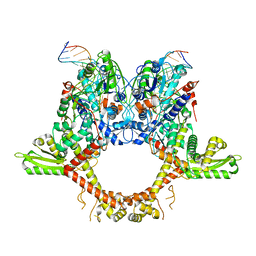 | |
6RKU
 
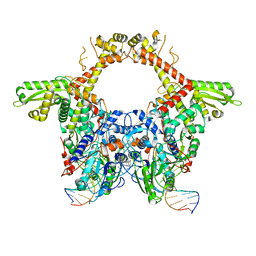 | | E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA Strand 1, DNA Strand 2, ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
4BZF
 
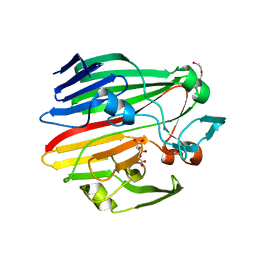 | | Crystal structure of galactose mutarotase GalM from Bacillus subtilis with trehalose | | Descriptor: | ACETATE ION, ALDOSE 1-EPIMERASE, CITRIC ACID, ... | | Authors: | Vanden Broeck, A, Sauvage, E, Herman, R, Kerff, F, Duez, C, Charlier, P. | | Deposit date: | 2013-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Galactose Mutarotase Galm from Bacillus Subtilis with Trehalose
To be Published
|
|
4BZG
 
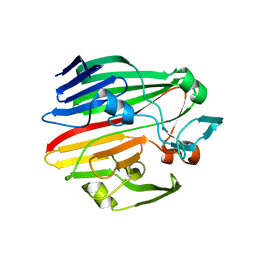 | | Crystal structure of galactose mutarotase GalM from Bacillus subtilis in complex with maltose | | Descriptor: | ALDOSE 1-EPIMERASE, CITRIC ACID, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Vanden Broeck, A, Sauvage, E, Herman, R, Kerff, F, Duez, C, Charlier, P. | | Deposit date: | 2013-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of Galactose Mutarotase Galm from Bacillus Subtilis in Complex with Maltose
To be Published
|
|
4BZH
 
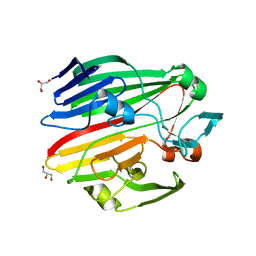 | | Crystal structure of galactose mutarotase GalM from Bacillus subtilis in complex with maltose and trehalose | | Descriptor: | ALDOSE 1-EPIMERASE, CITRIC ACID, GLYCEROL, ... | | Authors: | Vanden Broeck, A, Sauvage, E, Herman, R, Kerff, F, Duez, C, Charlier, P. | | Deposit date: | 2013-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Galactose Mutarotase Galm from Bacillus Subtilis in Complex with Maltose and Trehalose
To be Published
|
|
4BZE
 
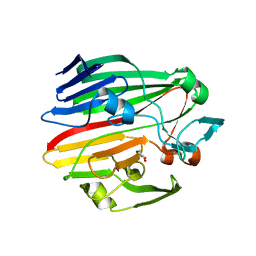 | | Crystal structure of galactose mutarotase GalM from Bacillus subtilis | | Descriptor: | ALDOSE 1-EPIMERASE, CITRIC ACID, GLYCEROL | | Authors: | Vanden Broeck, A, Sauvage, E, Herman, R, Kerff, F, Duez, C, Charlier, P. | | Deposit date: | 2013-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Galactose Mutarotase Galm from Bacillus Subtilis
To be Published
|
|
6ZY7
 
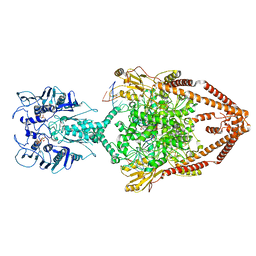 | | Cryo-EM structure of the entire Human topoisomerase II alpha in State 1 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.64 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
6ZY8
 
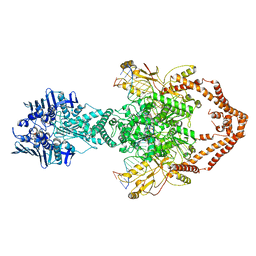 | | Cryo-EM structure of the entire Human topoisomerase II alpha in State 2 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
6ZY6
 
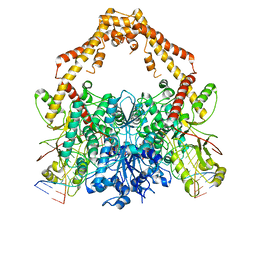 | | Cryo-EM structure of the Human topoisomerase II alpha DNA-binding/cleavage domain in State 2 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
6ZY5
 
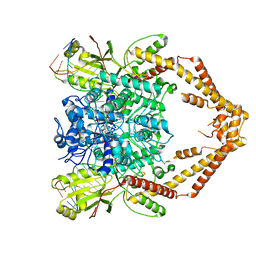 | | Cryo-EM structure of the Human topoisomerase II alpha DNA-binding/cleavage domain in State 1 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
8FLF
 
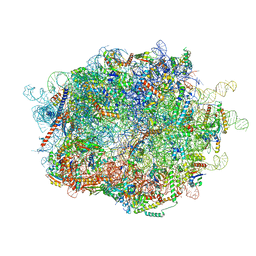 | |
8FKU
 
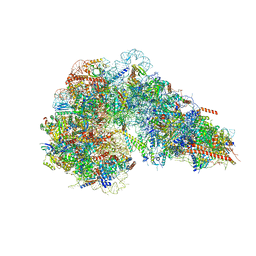 | |
8FLC
 
 | |
8FLD
 
 | |
8FKS
 
 | |
8FLA
 
 | |
8FKZ
 
 | |
8FL7
 
 | |
8FL3
 
 | |
8FLB
 
 | |
8FKW
 
 | |
