7N8I
 
 | |
7N8H
 
 | |
3BJE
 
 | |
5IFU
 
 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with Glyburide | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, CHLORIDE ION, ... | | Authors: | Dranow, D.M, Hewitt, S.N, Abendroth, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-26 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Biochemical and Structural Characterization of Selective Allosteric Inhibitors of the Plasmodium falciparum Drug Target, Prolyl-tRNA-synthetase.
ACS Infect Dis, 3, 2017
|
|
5IRD
 
 | |
5K0T
 
 | |
5K0S
 
 | |
5ELN
 
 | |
5ELO
 
 | |
4M97
 
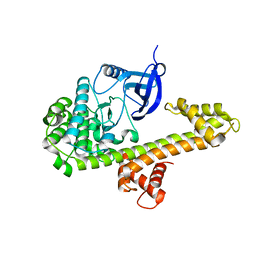 | | Calcium-Dependent Protein Kinase 1 from Neospora caninum | | Descriptor: | Calmodulin-like domain protein kinase isoenzyme gamma, related | | Authors: | Merritt, E.A. | | Deposit date: | 2013-08-14 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Neospora caninum Calcium-Dependent Protein Kinase 1 Is an Effective Drug Target for Neosporosis Therapy.
Plos One, 9, 2014
|
|
4MXA
 
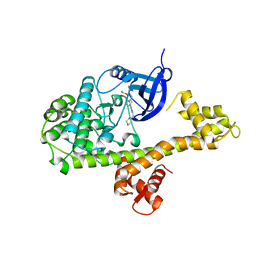 | | CDPK1 from Neospora caninum in complex with inhibitor RM-1-132 | | Descriptor: | 3-(6-ethoxynaphthalen-2-yl)-1-(piperidin-4-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-like domain protein kinase isoenzyme gamma, related | | Authors: | Merritt, E.A. | | Deposit date: | 2013-09-26 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Neospora caninum Calcium-Dependent Protein Kinase 1 Is an Effective Drug Target for Neosporosis Therapy.
Plos One, 9, 2014
|
|
4MX9
 
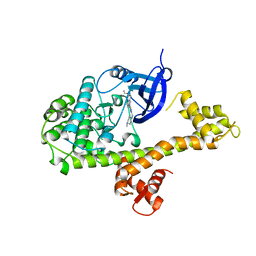 | | CDPK1 from Neospora caninum in complex with inhibitor UW1294 | | Descriptor: | 3-(6-ethoxynaphthalen-2-yl)-1-[(1-methylpiperidin-4-yl)methyl]-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-like domain protein kinase isoenzyme gamma, related | | Authors: | Merritt, E.A. | | Deposit date: | 2013-09-26 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Neospora caninum Calcium-Dependent Protein Kinase 1 Is an Effective Drug Target for Neosporosis Therapy.
Plos One, 9, 2014
|
|
4JBV
 
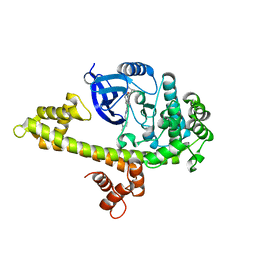 | |
8CUN
 
 | |
8CWA
 
 | |
8CTO
 
 | |
8DYA
 
 | |
4YSM
 
 | |
4YUQ
 
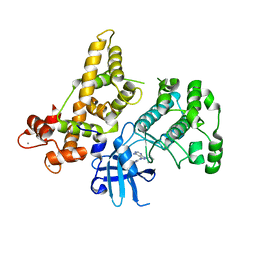 | | CDPK1 from Eimeria tenella in complex with inhibitor UW1354 | | Descriptor: | 1-cyclopentyl-3-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALCIUM ION, Calmodulin-like domain protein kinase | | Authors: | Merritt, E.A. | | Deposit date: | 2015-03-19 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CDPK is a druggable target in the apicomplexan parasite Eimeria
to be published
|
|
4YZB
 
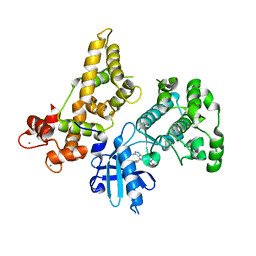 | | CDPK1 from Eimeria tenella in complex with inhibitor UW1521 | | Descriptor: | 4-(6-ethoxynaphthalen-2-yl)-6-(piperazin-1-ylmethyl)-2H-indazol-3-amine, CALCIUM ION, Calmodulin-like domain protein kinase | | Authors: | Merritt, E.A. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CDPK is a druggable target in the apicomplexan parasite Eimeria
to be published
|
|
7SOF
 
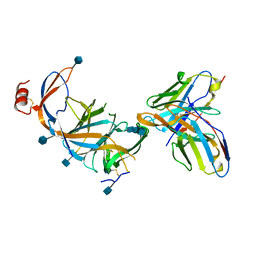 | |
7SOE
 
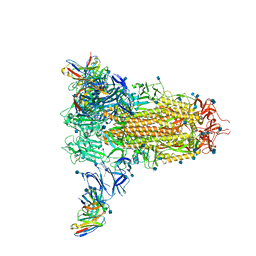 | |
7SOC
 
 | |
7SOB
 
 | |
7SO9
 
 | |
