2W62
 
 | | Saccharomyces cerevisiae Gas2p in complex with laminaripentaose | | Descriptor: | 1,4-BUTANEDIOL, GLYCOLIPID-ANCHORED SURFACE PROTEIN 2, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Schuettelkopf, A.W, Hurtado-Guerrero, R, van Aalten, D.M.F. | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Mechanisms of Yeast Cell Wall Glucan Remodeling.
J.Biol.Chem., 284, 2009
|
|
2VYO
 
 | | Chitin deacetylase family member from Encephalitozoon cuniculi | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Urch, J.E, Hurtado-Guerrero, R, Texier, C, Van Aalten, D.M.F. | | Deposit date: | 2008-07-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of a Putative Polysaccharide Deacetylase of the Human Parasite Encephalitozoon Cuniculi.
Protein Sci., 18, 2009
|
|
2VSY
 
 | | Xanthomonas campestris putative OGT (XCC0866), apostructure | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schuettelkopf, A.W, Clarke, A.J, van Aalten, D.M.F. | | Deposit date: | 2008-05-01 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights Into Mechanism and Specificity of O-Glcnac Transferase.
Embo J., 27, 2008
|
|
2W3Z
 
 | | Structure of a Streptococcus mutans CE4 esterase | | Descriptor: | PHOSPHATE ION, PUTATIVE DEACETYLASE, ZINC ION | | Authors: | Deng, D.M, Urch, J.E, ten Cate, J.M, Rao, V.A, van Aalten, D.M.F, Crielaard, W. | | Deposit date: | 2008-11-17 | | Release date: | 2008-12-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Streptococcus Mutans Smu.623C Codes for a Functional, Metal Dependent Polysaccharide Deacetylase that Modulates Interactions with Salivary Agglutinin.
J.Bacteriol., 191, 2009
|
|
2WZF
 
 | | Legionella pneumophila glucosyltransferase crystal structure | | Descriptor: | GLUCOSYLTRANSFERASE, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Hurtado-Guerrero, R, Zusman, T, Pathak, S, Ibrahim, A.F.M, Shepherd, S, Prescott, A, Segal, G, van Aalten, D.M.F. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Mechanism of Elongation Factor 1A Inhibition by a Legionella Pneumophila Glycosyltransferase.
Biochem.J., 426, 2010
|
|
4EZA
 
 | | Crystal structure of the atypical phosphoinositide (aPI) binding domain of IQGAP2 | | Descriptor: | Ras GTPase-activating-like protein IQGAP2 | | Authors: | Van Aalten, D.M.F, Dixon, M.J, Gray, A, Schenning, M, Agacan, M, Leslie, N.R, Downes, C.P, Batty, I.H, Nedyalkova, L, Tempel, W, Tong, Y, Zhong, N, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-02 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | IQGAP Proteins Reveal an Atypical Phosphoinositide (aPI) Binding Domain with a Pseudo C2 Domain Fold.
J.Biol.Chem., 287, 2012
|
|
1CW9
 
 | | DNA DECAMER WITH AN ENGINEERED CROSSLINK IN THE MINOR GROOVE | | Descriptor: | 5'-D(*CP*CP*AP*GP*(G47)P*CP*CP*TP*GP*G)-3', CALCIUM ION | | Authors: | van Aalten, D.M.F, Erlanson, D.A, Verdine, G.L, Joshua-Tor, L. | | Deposit date: | 1999-08-26 | | Release date: | 1999-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A structural snapshot of base-pair opening in DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1D7E
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 1999-10-17 | | Release date: | 2000-03-31 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Conformational substates in different crystal forms of the photoactive yellow protein--correlation with theoretical and experimental flexibility.
Protein Sci., 9, 2000
|
|
1HBK
 
 | | Acyl-CoA binding protein from Plasmodium falciparum | | Descriptor: | ACYL-COA BINDING PROTEIN, COENZYME A, MYRISTIC ACID, ... | | Authors: | van Aalten, D.M.F. | | Deposit date: | 2001-04-16 | | Release date: | 2001-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding site differences revealed by crystal structures of Plasmodium falciparum and bovine acyl-CoA binding protein.
J. Mol. Biol., 309, 2001
|
|
1KOU
 
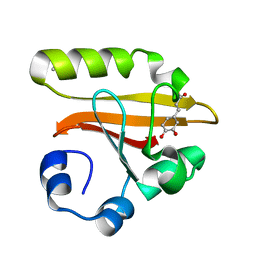 | | Crystal Structure of the Photoactive Yellow Protein Reconstituted with Caffeic Acid at 1.16 A Resolution | | Descriptor: | CAFFEIC ACID, N-BUTANE, PHOTOACTIVE YELLOW PROTEIN | | Authors: | van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2001-12-22 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the photoactive yellow protein reconstituted with caffeic acid at 1.16 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1HB8
 
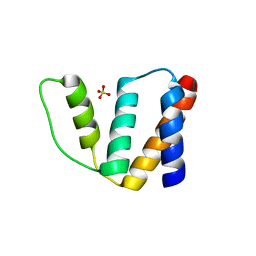 | | Structure of bovine Acyl-CoA binding protein in tetragonal crystal form | | Descriptor: | ACYL-COA BINDING PROTEIN, SULFATE ION | | Authors: | Zou, J.Y, Kleywegt, G.J, Bergfors, T, Knudsen, J, Jones, T.A. | | Deposit date: | 2001-04-12 | | Release date: | 2002-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding Site Differences Revealed by Crystal Structures of Plasmodium Falciparum and Bovine Acyl-Coa Binding Protein
J.Mol.Biol., 309, 2001
|
|
1HB6
 
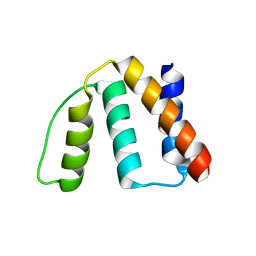 | | Structure of bovine Acyl-CoA binding protein in orthorhombic crystal form | | Descriptor: | ACYL-COA BINDING PROTEIN, CADMIUM ION | | Authors: | Zou, J.Y, Kleywegt, G.J, Bergfors, T, Knudsen, J, Jones, T.A. | | Deposit date: | 2001-04-12 | | Release date: | 2002-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding Site Differences Revealed by Crystal Structures of Plasmodium Falciparum and Bovine Acyl-Coa Binding Protein
J.Mol.Biol., 309, 2001
|
|
4V11
 
 | | Structure of Synaptotagmin-1 with SV2A peptide phosphorylated at Thr84 | | Descriptor: | CALCIUM ION, GLYCEROL, SYNAPTIC VESICLE GLYCOPROTEIN 2A, ... | | Authors: | Zhang, N, Gordon, S.L, Fritsch, M.J, Esoof, N, Campbell, D, Gourlay, R, Velupillai, S, Macartney, T, Peggie, M, vanAalten, D.M.F, Cousin, M.A, Alessi, D.R. | | Deposit date: | 2014-09-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphorylation of Synaptic Vesicle Protein 2A at Thr84 by Casein Kinase 1 Family Kinases Controls the Specific Retrieval of Synaptotagmin-1.
J.Neurosci., 35, 2015
|
|
6G9W
 
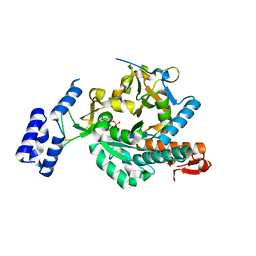 | | Crystal Structure of Aspergillus fumigatus UDP-N-acetylglucosamine pyrophosphorylase in complex with UTP | | Descriptor: | UDP-N-acetylglucosamine pyrophosphorylase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Raimi, O.G, Hurtado-Guerrero, R, Borodin, V, Urbaniak, M, Ferguson, M, van Aalten, D. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A mechanism-inspired UDP-N-acetylglucosamine pyrophosphorylase inhibitor
RSC Chem. Biol., 2020
|
|
7U34
 
 | | The structure of phosphoglucose isomerase from Aspergillus fumigatus | | Descriptor: | CHLORIDE ION, CITRATE ANION, GLYCEROL, ... | | Authors: | Yan, K, Kowalski, B, Fang, W, van Aalten, D. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Phosphoglucose Isomerase Is Important for Aspergillus fumigatus Cell Wall Biogenesis.
Mbio, 13, 2022
|
|
6TDF
 
 | | Crystal structure of Aspergillus fumigatus Glucosamine-6-phosphate N-acetyltransferase 1 in complex with compound 3 | | Descriptor: | 2-[[3,5-bis(chloranyl)-4-(4~{H}-1,2,4-triazol-3-yl)phenyl]-(2-hydroxyethyl)amino]ethanol, 6-O-phosphono-alpha-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Raimi, O.G, Stanley, M, Lockhart, D. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Targeting a critical step in fungal hexosamine biosynthesis.
J.Biol.Chem., 295, 2020
|
|
6TN3
 
 | |
1W17
 
 | |
6G9V
 
 | | Crystal structure of Aspergillus fumigatus UDP-N-acetylglucosamine pyrophosphorylase(AfUAP1) in complex with UDPGlcNAc, pyrophosphate and Mg2+ | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, UDP-N-acetylglucosamine pyrophosphorylase, ... | | Authors: | Raimi, O.G, Hurtado-Guerrero, R, Borodin, V, Urbaniak, M, Ferguson, M, van Aalten, D. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A mechanism-inspired UDP-N-acetylglucosamine pyrophosphorylase inhibitor
RSC Chem. Biol., 2020
|
|
4WPG
 
 | | Group A Streptococcus GacA is an essential dTDP-4-dehydrorhamnose reductase (RmlD) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, ... | | Authors: | van der Beek, S.L, Le Breton, Y, Ferenbach, A.T, van Aalten, D.M.F, Navratilova, I, McIver, K, van Sorge, N.M, Dorfmueller, H.C. | | Deposit date: | 2014-10-18 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | GacA is essential for Group A Streptococcus and defines a new class of monomeric dTDP-4-dehydrorhamnose reductases (RmlD).
Mol.Microbiol., 98, 2015
|
|
5LWV
 
 | | Human OGT in complex with UDP and fused substrate peptide (HCF1) | | Descriptor: | GLYCEROL, Host cell factor 1,UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, PHOSPHATE ION, ... | | Authors: | Raimi, O, Rafie, K, Kapuria, V, Herr, W, van Aalten, D. | | Deposit date: | 2016-09-19 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats.
Open Biol, 7, 2017
|
|
4XI9
 
 | | Human OGT in complex with UDP-5S-GlcNAc and substrate peptide (RBL2) | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Retinoblastoma-like protein 2, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Schimpl, M, van Aalten, D.M.F. | | Deposit date: | 2015-01-06 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XIF
 
 | | Human OGT in complex with UDP-5S-GlcNAc and substrate peptide (keratin-7) | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Keratin, type II cytoskeletal 7, ... | | Authors: | Schimpl, M, van Aalten, D.M.F. | | Deposit date: | 2015-01-06 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5DIY
 
 | | Thermobaculum terrenum O-GlcNAc hydrolase mutant - D120N | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hyaluronidase, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 | | Authors: | Ostrowski, A, Gundogdu, M, Ferenbach, A.T, Lebedev, A, van Aalten, D.M.F. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Evidence for a Functional O-Linked N-Acetylglucosamine (O-GlcNAc) System in the Thermophilic Bacterium Thermobaculum terrenum.
J.Biol.Chem., 290, 2015
|
|
5FIH
 
 | | SACCHAROMYCES CEREVISIAE GAS2P (E176Q MUTANT) IN COMPLEX WITH LAMINARITETRAOSE AND LAMINARIPENTAOSE | | Descriptor: | 1,3-BETA-GLUCANOSYLTRANSFERASE, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Raich, L, Borodkin, V, van Aalten, D.M.F, Hurtado-Guerrero, R, Rovira, C. | | Deposit date: | 2015-09-25 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Trapped Covalent Intermediate of a Glycoside Hydrolase on the Pathway to Transglycosylation. Insights from Experiments and Quantum Mechanics/Molecular Mechanics Simulations.
J. Am. Chem. Soc., 138, 2016
|
|
