6RU6
 
 | | Crystal structure of Casein Kinase I delta (CK1d) in complex with monophosphorylated p63 PAD1P peptide | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase I isoform delta, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Chaikuad, A, Tuppi, M, Gebel, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | p63 uses a switch-like mechanism to set the threshold for induction of apoptosis.
Nat.Chem.Biol., 16, 2020
|
|
6RU7
 
 | | Crystal structure of Casein Kinase I delta (CK1d) in complex with double phosphorylated p63 PAD2P peptide | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Tuppi, M, Gebel, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | p63 uses a switch-like mechanism to set the threshold for induction of apoptosis.
Nat.Chem.Biol., 16, 2020
|
|
6RU8
 
 | | Crystal structure of Casein Kinase I delta (CK1d) in complex with triple phosphorylated p63 PAD3P peptide | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Tuppi, M, Gebel, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | p63 uses a switch-like mechanism to set the threshold for induction of apoptosis.
Nat.Chem.Biol., 16, 2020
|
|
8P9C
 
 | | Crystal structure of p63-p73 heterotetramer (tetramerisation domain) in complex with darpin 1810 F11 | | Descriptor: | 1,2-ETHANEDIOL, Darpin 1810 F11, Tumor protein 63, ... | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | DARPins detect the formation of hetero-tetramers of p63 and p73 in epithelial tissues and in squamous cell carcinoma.
Cell Death Dis, 14, 2023
|
|
8P9D
 
 | | Crystal structure of p63-p73 heterotetramer (tetramerisation domain) in complex with darpin 1810 A2 | | Descriptor: | Darpin 1810 A2, Tumor protein 63, Tumor protein p73 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | DARPins detect the formation of hetero-tetramers of p63 and p73 in epithelial tissues and in squamous cell carcinoma.
Cell Death Dis, 14, 2023
|
|
8P9E
 
 | | Crystal structure of wild type p63-p73 heterotetramer (tetramerisation domain) in complex with darpin 1810 F11 | | Descriptor: | Darpin 1810 F11, GLYCEROL, Isoform 2 of Tumor protein 63, ... | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | DARPins detect the formation of hetero-tetramers of p63 and p73 in epithelial tissues and in squamous cell carcinoma.
Cell Death Dis, 14, 2023
|
|
6FGS
 
 | | Solution structure of p300Taz2-p73TA1 | | Descriptor: | Histone acetyltransferase p300,Tumor protein p73, ZINC ION | | Authors: | Gebel, J, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Regulation of the Activity in the p53 Family Depends on the Organization of the Transactivation Domain.
Structure, 26, 2018
|
|
6FGN
 
 | | Solution Structure of p300Taz2-p63TA | | Descriptor: | Histone acetyltransferase p300,Tumor protein 63, ZINC ION | | Authors: | Gebel, J, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Regulation of the Activity in the p53 Family Depends on the Organization of the Transactivation Domain.
Structure, 26, 2018
|
|
8RJW
 
 | | Human RAD52 open ring - ssDNA complex | | Descriptor: | DNA repair protein RAD52 homolog, MAGNESIUM ION, ssDNA | | Authors: | Liang, C.C, West, S.C. | | Deposit date: | 2023-12-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Mechanism of single-stranded DNA annealing by RAD52-RPA complex.
Nature, 629, 2024
|
|
8RK2
 
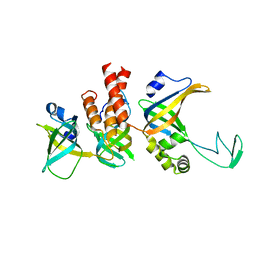 | | Human Replication protein A (RPA; trimeric core) - ssDNA complex | | Descriptor: | Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit, Replication protein A 70 kDa DNA-binding subunit, ... | | Authors: | Liang, C.C, West, S.C. | | Deposit date: | 2023-12-22 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of single-stranded DNA annealing by RAD52-RPA complex.
Nature, 629, 2024
|
|
8RJ3
 
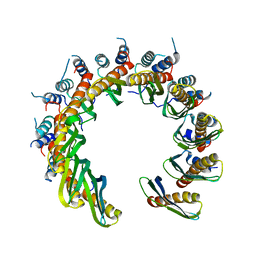 | | Human RAD52 open ring conformation | | Descriptor: | DNA repair protein RAD52 homolog | | Authors: | Liang, C.C, West, S.C. | | Deposit date: | 2023-12-19 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of single-stranded DNA annealing by RAD52-RPA complex.
Nature, 629, 2024
|
|
8RIL
 
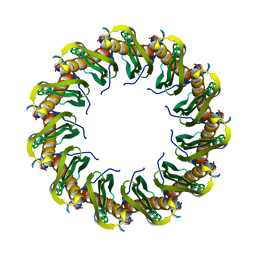 | | Human RAD52 closed ring conformation | | Descriptor: | DNA repair protein RAD52 homolog | | Authors: | Liang, C.C, West, S.C. | | Deposit date: | 2023-12-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of single-stranded DNA annealing by RAD52-RPA complex.
Nature, 629, 2024
|
|
7Z7E
 
 | | Crystal structure of p63 DNA binding domain in complex with inhibitory DARPin G4 | | Descriptor: | DARPIN, Isoform 4 of Tumor protein 63, ZINC ION | | Authors: | Strubel, A, Gebel, J, Chaikuad, A, Muenick, P, Doetsch, V. | | Deposit date: | 2022-03-15 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
7Z71
 
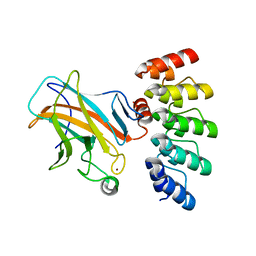 | | Crystal structure of p63 DBD in complex with darpin C14 | | Descriptor: | Darpin C14, Isoform 4 of Tumor protein 63, ZINC ION | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
7Z73
 
 | | Crystal structure of p63 tetramerization domain in complex with darpin 8F1 | | Descriptor: | Darpin 8F1, Isoform 2 of Tumor protein 63 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
7Z72
 
 | | Crystal structure of p63 SAM in complex with darpin A5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Darpin A5, Isoform 9 of Tumor protein 63 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
