2Z9S
 
 | | Crystal Structure Analysis of rat HBP23/Peroxiredoxin I, Cys52Ser mutant | | Descriptor: | Peroxiredoxin-1 | | Authors: | Matsumura, T, Okamoto, K, Nishino, T, Abe, Y. | | Deposit date: | 2007-09-25 | | Release date: | 2007-11-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dimer-Oligomer Interconversion of Wild-type and Mutant Rat 2-Cys Peroxiredoxin: DISULFIDE FORMATION AT DIMER-DIMER INTERFACES IS NOT ESSENTIAL FOR DECAMERIZATION
J.Biol.Chem., 283, 2008
|
|
6KRO
 
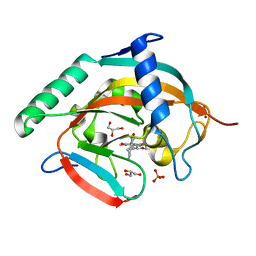 | | Tankyrase-2 in complex with RK-582 | | Descriptor: | 6-[(2S,6R)-2,6-dimethylmorpholin-4-yl]-4-fluoranyl-1-methyl-1'-(8-methyl-4-oxidanylidene-3,5,6,7-tetrahydropyrido[2,3-d]pyrimidin-2-yl)spiro[indole-3,4'-piperidine]-2-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2019-08-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Discovery of an Orally Efficacious Spiroindolinone-Based Tankyrase Inhibitor for the Treatment of Colon Cancer.
J.Med.Chem., 63, 2020
|
|
5ZQQ
 
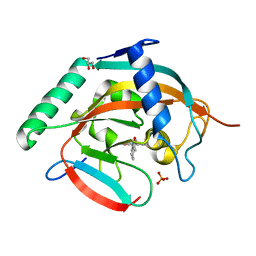 | | Tankyrase-2 in complex with compound 52 | | Descriptor: | 1-methyl-1'-(4-oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)spiro[indole-3,4'-piperidin]-2(1H)-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
5ZQO
 
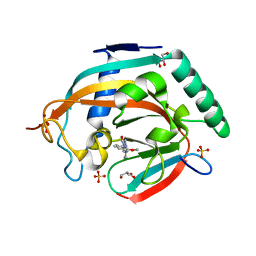 | | Tankyrase-2 in complex with compound 1a | | Descriptor: | 2-[4-(2-methoxyphenyl)piperazin-1-yl]-5,6,7,8-tetrahydroquinazolin-4(3H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
5ZQP
 
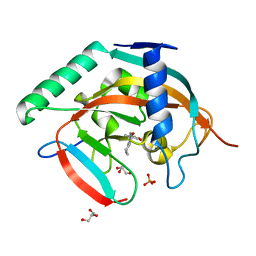 | | Tankyrase-2 in complex with compound 12 | | Descriptor: | 1'-(4-oxo-3,4,5,6,7,8-hexahydroquinazolin-2-yl)-2H-spiro[1-benzofuran-3,4'-piperidin]-2-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
5ZQR
 
 | | Tankyrase-2 in complex with compound 40c | | Descriptor: | 2-[4,6-difluoro-1-(2-hydroxyethyl)-1,2-dihydro-1'H-spiro[indole-3,4'-piperidin]-1'-yl]-5,6,7,8-tetrahydroquinazolin-4(3H)-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-04-19 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
6A84
 
 | | Tankyrase-2 in complex with compound 15d | | Descriptor: | 2-(4-chloro-1,2-dihydro-1'H-spiro[indole-3,4'-piperidin]-1'-yl)-5,6,7,8-tetrahydroquinazolin-4(3H)-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-07-06 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
6IGU
 
 | | Crystal structure of the hydrolytic antibody Fab 9C10 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IMMUNOGLOBULIN 9C10 H CHAIN, IMMUNOGLOBULIN 9C10 L CHAIN, ... | | Authors: | Yamaguchi, A, Tada, T, Tsuchiya, Y, Tsumuraya, T, Fujii, I. | | Deposit date: | 2018-09-26 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of the complex of the hydrolytic antibody Fab 9C10 and a transition-state analog
To Be Published
|
|
4YTZ
 
 | | Rat xanthine oxidoreductase, C-terminal deletion protein variant, crystal grown without dithiothreitol | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase
Febs J., 282, 2015
|
|
4YTY
 
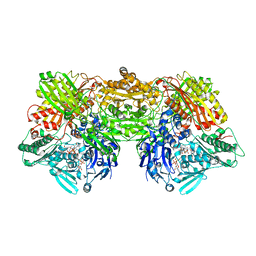 | | Structure of rat xanthine oxidoreductase, C535A/C992R/C1324S, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase.
Febs J., 282, 2015
|
|
4YSW
 
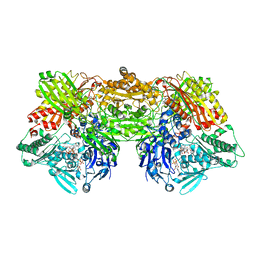 | | Structure of rat xanthine oxidoreductase, C-terminal deletion protein variant, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F. | | Deposit date: | 2015-03-17 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase.
Febs J., 282, 2015
|
|
6A9K
 
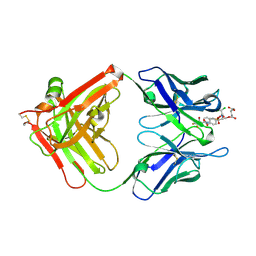 | | Crystal structure of the complex of the hydrolytic antibody Fab 9C10 with a transition-state analog | | Descriptor: | 5-[(2R,3R)-2-[2,2-bis(chloranyl)ethanoylamino]-3-(4-nitrophenyl)-3-[oxidanyl-[[4-[2,2,2-tris(fluoranyl)ethanoylamino]phenyl]methyl]phosphoryl]oxy-propoxy]-5-oxidanylidene-pentanoic acid, IMMUNOGLOBULIN 9C10 H CHAIN, IMMUNOGLOBULIN 9C10 L CHAIN | | Authors: | Tsuchiya, Y, Fujii, I, Tada, T, Yamaguchi, A, Tsumuraya, T, Kumon, A. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of the hydrolytic antibody Fab 9C10 with a transition-state analog
To Be Published
|
|
1WYG
 
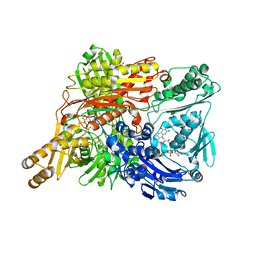 | | Crystal Structure of a Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S) | | Descriptor: | 2-HYDROXYBENZOIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Hori, H, Matsumura, T, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2005-02-14 | | Release date: | 2005-05-31 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of the Conversion of Xanthine Dehydrogenase to Xanthine Oxidase: IDENTIFICATION OF THE TWO CYSTEINE DISULFIDE BONDS AND CRYSTAL STRUCTURE OF A NON-CONVERTIBLE RAT LIVER XANTHINE DEHYDROGENASE MUTANT
J.Biol.Chem., 280, 2005
|
|
2P6V
 
 | | Structure of TAFH domain of the human TAF4 subunit of TFIID | | Descriptor: | SULFATE ION, Transcription initiation factor TFIID subunit 4 | | Authors: | Wang, X, Truckses, D.M, Takada, S, Matsumura, T, Tanese, N, Jacobson, R.H. | | Deposit date: | 2007-03-19 | | Release date: | 2007-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conserved region I of human coactivator TAF4 binds to a short hydrophobic motif present in transcriptional regulators.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5XQW
 
 | | Catalytic antibody 7B9 | | Descriptor: | Fab fragment of catalytic antibody 7B9, heavy chain, light chain, ... | | Authors: | Ito, N, Fujii, I, Tsumuraya, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the broad substrate tolerance of the antibody 7B9-catalyzed hydrolysis of p-nitrobenzyl esters.
Bioorg. Med. Chem., 26, 2018
|
|
1CT8
 
 | | CATALYTIC ANTIBODY 7C8 COMPLEX | | Descriptor: | 7C8 FAB FRAGMENT; LONG CHAIN, 7C8 FAB FRAGMENT; SHORT CHAIN, SULFATE ION, ... | | Authors: | Gigant, B, Tsumuraya, T, Fujii, I, Knossow, M. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Diverse structural solutions to catalysis in a family of antibodies.
Structure Fold.Des., 7, 1999
|
|
2E3T
 
 | | Crystal structure of rat xanthine oxidoreductase mutant (W335A and F336L) | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Asai, R, Nishino, T, Matsumura, T, Okamoto, K, Pai, E.F, Nishino, T. | | Deposit date: | 2006-11-28 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Two mutations convert mammalian xanthine oxidoreductase to highly superoxide-productive xanthine oxidase
J.Biochem.(Tokyo), 141, 2007
|
|
2E1Q
 
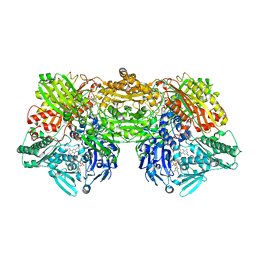 | | Crystal Structure of Human Xanthine Oxidoreductase mutant, Glu803Val | | Descriptor: | 2-HYDROXYBENZOIC ACID, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Yamaguchi, Y, Matsumura, T, Ichida, K, Okamoto, K, Nishino, T. | | Deposit date: | 2006-10-27 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human xanthine oxidase changes its substrate specificity to aldehyde oxidase type upon mutation of amino acid residues in the active site: roles of active site residues in binding and activation of purine substrate
J.Biochem.(Tokyo), 141, 2007
|
|
3WIN
 
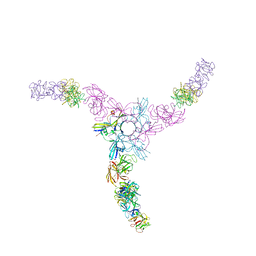 | | Clostridium botulinum Hemagglutinin | | Descriptor: | 17 kD hemagglutinin component, HA1, HA3 | | Authors: | Amatsu, S, Sugawara, Y, Matsumura, T, Fujinaga, Y, Kitadokoro, K. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of Clostridium botulinum Whole Hemagglutinin Reveals a Huge Triskelion-shaped Molecular Complex
J.Biol.Chem., 288, 2013
|
|
2DTM
 
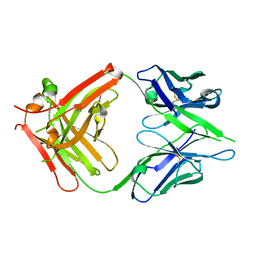 | | Thermodynamic and structural analyses of hydrolytic mechanism by catalytic antibodies | | Descriptor: | IMMUNOGLOBULIN 6D9 | | Authors: | Oda, M, Ito, N, Tsumuraya, T, Suzuki, K, Fujii, I. | | Deposit date: | 2006-07-13 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
7EA6
 
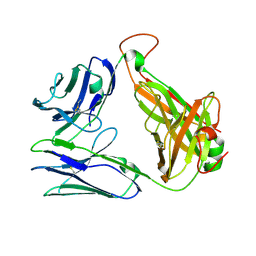 | | Crystal structure of TCR-017 ectodomain | | Descriptor: | T cell receptor 017 alpha chain, T cell receptor 017 beta chain | | Authors: | Nagae, M, Yamasaki, S. | | Deposit date: | 2021-03-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18000245 Å) | | Cite: | Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients.
J.Exp.Med., 218, 2021
|
|
4YRW
 
 | | rat xanthine oxidoreductase, C-terminal deletion protein variant | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K. | | Deposit date: | 2015-03-16 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase
Febs J., 282, 2015
|
|
2DQU
 
 | | Crystal form II: high resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog | | Descriptor: | IMMUNOGLOBULIN 6D9, [1-(3-DIMETHYLAMINO-PROPYL)-3-ETHYL-UREIDO]-[4-(2,2,2-TRIFLUORO-ACETYLAMINO)-BENZYL]PHOSPHINIC ACID-2-(2,2-DIHYDRO-ACETYLAMINO)-3-HYDROXY-1-(4-NITROPHENYL)-PROPYL ESTER | | Authors: | Kristensen, O, Vassylyev, D.G, Tanaka, F, Ito, N, Morikawa, K, Fujii, I. | | Deposit date: | 2006-05-30 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
2DQT
 
 | | High resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog | | Descriptor: | IMMUNOGLOBULIN 6D9, [1-(3-DIMETHYLAMINO-PROPYL)-3-ETHYL-UREIDO]-[4-(2,2,2-TRIFLUORO-ACETYLAMINO)-BENZYL]PHOSPHINIC ACID-2-(2,2-DIHYDRO-ACETYLAMINO)-3-HYDROXY-1-(4-NITROPHENYL)-PROPYL ESTER | | Authors: | Kristensen, O, Vassylyev, D.G, Tanaka, F, Ito, N, Morikawa, K, Fujii, I. | | Deposit date: | 2006-05-30 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
6K9U
 
 | | Discovery of Pyrazolo[1,5-a]pyrimidine Derivative as a Highly Selective PDE10A Inhibitor | | Descriptor: | 2-(3,7-dimethylquinoxalin-2-yl)-~{N}-(oxan-4-yl)-5-pyrrolidin-1-yl-pyrazolo[1,5-a]pyrimidin-7-amine, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Takedomi, K, Koizumi, Y. | | Deposit date: | 2019-06-18 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of a pyrazolo[1,5-a]pyrimidine derivative (MT-3014) as a highly selective PDE10A inhibitor via core structure transformation from the stilbene moiety.
Bioorg.Med.Chem., 27, 2019
|
|
