2FE3
 
 | |
5VFY
 
 | | Structure of an accessory protein of the pCW3 relaxosome | | Descriptor: | TcpK | | Authors: | Traore, D.A.K, Wisniewski, J.A, Flanigan, S.F, Conroy, P.J, Panjikar, S, Mok, Y.-F, Lao, C, Griffin, M.D.W, Adams, V, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.
Nat Commun, 9, 2018
|
|
5VFX
 
 | | Structure of an accessory protein of the pCW3 relaxosome in complex with the origin of transfer (oriT) DNA | | Descriptor: | TcpK, oriT | | Authors: | Traore, D.A.K, Wisniewski, J.A, Flanigan, S.F, Conroy, P.J, Panjikar, S, Mok, Y.-F, Lao, C, Griffin, M.D.W, Adams, V, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.
Nat Commun, 9, 2018
|
|
6O1Z
 
 | | Structure of pCW3 conjugation coupling protein TcpA hexagonal crystal form | | Descriptor: | DNA translocase coupling protein | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
6O1Y
 
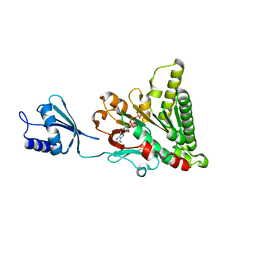 | | Structure of pCW3 conjugation coupling protein TcpA monomeric form with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA translocase coupling protein, beta-D-glucopyranose | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
6O1W
 
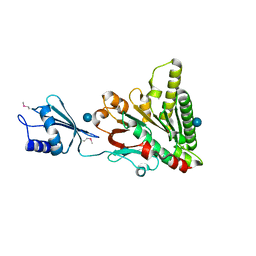 | | Structure of pCW3 conjugation coupling protein TcpA monomer orthorhombic crystal form | | Descriptor: | DNA translocase coupling protein, beta-D-glucopyranose | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
6O1X
 
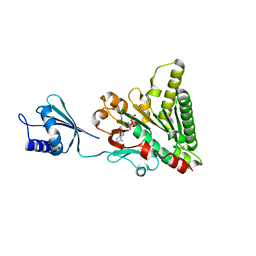 | | Structure of pCW3 conjugation coupling protein TcpA monomer form with ATPgS | | Descriptor: | DNA translocase coupling protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, beta-D-glucopyranose | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
3F8N
 
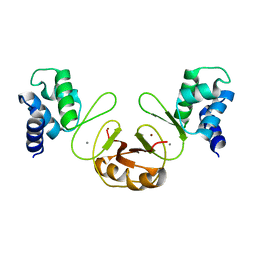 | | Crystal structure of PerR-Zn-Mn | | Descriptor: | MANGANESE (II) ION, Peroxide operon regulator, ZINC ION | | Authors: | Traore, D.A.K, Ferrer, J.-L, Jacquamet, L, Duarte, V, Latour, J.-M. | | Deposit date: | 2008-11-13 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding
Mol.Microbiol., 73, 2009
|
|
7BCW
 
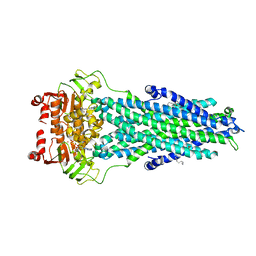 | | Structure of MsbA in Salipro with ADP vanadate | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-DIPHOSPHATE, ATP-dependent lipid A-core flippase, ... | | Authors: | Traore, D.A.K, Tidow, H. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of MsbA in saposin-lipid nanoparticles (Salipro) provides insights into nucleotide coordination.
Febs J., 289, 2022
|
|
3VKE
 
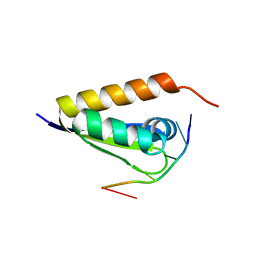 | |
2WCI
 
 | | Structure of E. coli monothiol glutaredoxin GRX4 homodimer | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLUTAREDOXIN-4, GLUTATHIONE, ... | | Authors: | Iwema, T, Picchiocci, A, Traore, D.A.K, Ferrer, J.-L, Chauvat, F, Jacquamet, L. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Delivery of the Intact [Fe2S2] Cluster by Monothiol Glutaredoxin.
Biochemistry, 48, 2009
|
|
2WHG
 
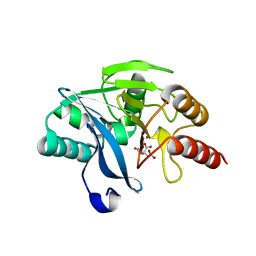 | | Crystal Structure of the Di-Zinc Metallo-beta-lactamase VIM-4 from Pseudomonas aeruginosa | | Descriptor: | CITRATE ANION, VIM-4 METALLO-BETA-LACTAMASE, ZINC ION | | Authors: | Lassaux, P, Traore, D.A.K, Galleni, M, Ferrer, J.L. | | Deposit date: | 2009-05-05 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Structural Characterization of the Subclass B1 Metallo-{Beta}-Lactamase Vim-4.
Antimicrob.Agents Chemother., 55, 2011
|
|
2RGV
 
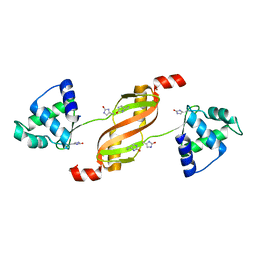 | |
3W56
 
 | | Structure of a C2 domain | | Descriptor: | C2 domain protein | | Authors: | Traore, D.A.K, Whisstock, J.C. | | Deposit date: | 2013-01-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Defining the interaction of perforin with calcium and the phospholipid membrane.
Biochem.J., 456, 2013
|
|
3W57
 
 | | Structure of a C2 domain | | Descriptor: | C2 domain protein, CALCIUM ION | | Authors: | Traore, D.A.K, Whisstock, J.C. | | Deposit date: | 2013-01-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Defining the interaction of perforin with calcium and the phospholipid membrane.
Biochem.J., 456, 2013
|
|
4V3N
 
 | | Membrane bound pleurotolysin prepore (TMH2 strand lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V2T
 
 | | Membrane embedded pleurotolysin pore with 13 fold symmetry | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
1ZTG
 
 | | human alpha polyC binding protein KH1 | | Descriptor: | 5'-D(P*CP*CP*CP*TP*CP*CP*CP*T)-3', POLY(RC)-BINDING PROTEIN 1 | | Authors: | Sidiqi, M, Wilce, J.A, Barker, A, Schmidgerger, J, Leedman, P.J, Wilce, M.C.J. | | Deposit date: | 2005-05-27 | | Release date: | 2006-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Contribution of the first K-homology domain of poly(C)-binding protein 1 to its affinity and specificity for C-rich oligonucleotides
Nucleic Acids Res., 40, 2012
|
|
4TZG
 
 | | Crystal structure of eCGP123, an extremely thermostable green fluorescent protein | | Descriptor: | Fluorescent Protein | | Authors: | Close, D.W, Don Paul, C, Traore, D.A.K, Wilce, M.C.J, Prescott, M, Bradbury, A.R.M. | | Deposit date: | 2014-07-10 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermal green protein, an extremely stable, nonaggregating fluorescent protein created by structure-guided surface engineering.
Proteins, 83, 2015
|
|
9FFH
 
 | | Native capsid of Rhodobacter microvirus Ebor computed with I4 symmetry | | Descriptor: | Major capsid protein | | Authors: | Bardy, P, MacDonald, C.I.W, Jenkins, H.T, Chechik, M, Hart, S.J, Turkenburg, J.P, Blaza, J.N, Fogg, P.C.M, Antson, A.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A stargate mechanism of Microviridae genome delivery unveiled by cryogenic electron tomography.
Biorxiv, 2024
|
|
9FFG
 
 | | Empty capsid of Rhodobacter microvirus Ebor computed with I4 symmetry | | Descriptor: | Major capsid protein | | Authors: | Bardy, P, MacDonald, C.I.W, Jenkins, H.T, Byrom, L, Chechik, M, Hart, S.J, Turkenburg, J.P, Blaza, J.N, Fogg, P.C.M, Antson, A.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A stargate mechanism of Microviridae genome delivery unveiled by cryogenic electron tomography.
Biorxiv, 2024
|
|
4PPL
 
 | | Crystal structure of eCGP123 H193Q variant at pH 7.5 | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
4PPJ
 
 | | Crystal structure of Phanta, a weakly fluorescent photochromic GFP-like protein. ON state | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
4PPK
 
 | | Crystal structure of eCGP123 T69V variant at pH 7.5 | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
3VIC
 
 | | Green-fluorescent variant of the non-fluorescent chromoprotein Rtms5 | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Traore, D.A.K, Byres, E, Wilce, M, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2011-09-28 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Green Fluorescent Protein Containing a QFG Tri-Peptide Chromophore: Optical Properties and X-Ray Crystal Structure.
Plos One, 7, 2012
|
|
