1N1C
 
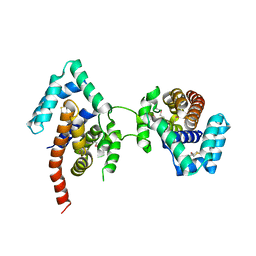 | | Crystal Structure Of The Dimeric TorD Chaperone From Shewanella Massilia | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, TorA specific chaperone | | Authors: | Tranier, S, Iobbi-Nivol, C, Mortier-Barriere, I, Birck, C, Mejean, V, Samama, J.-P. | | Deposit date: | 2002-10-17 | | Release date: | 2003-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Novel Protein Fold and Extreme Domain Swapping in the Dimeric TorD Chaperone from Shewanella massilia
Structure, 11, 2003
|
|
1E25
 
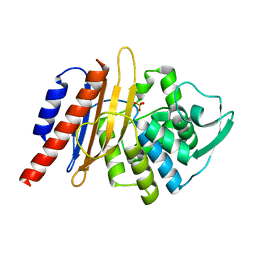 | | The high resolution structure of PER-1 class A beta-lactamase | | Descriptor: | EXTENDED-SPECTRUM BETA-LACTAMASE PER-1, SULFATE ION | | Authors: | Tranier, S, Bouthors, A.T, Maveyraud, L, Guillet, V, Sougakoff, W, Samama, J.P. | | Deposit date: | 2000-05-17 | | Release date: | 2000-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The High Resolution Crystal Structure for Class a Beta-Lactamase Per-1 Reveals the Bases for its Increase in Breadth of Activity
J.Biol.Chem., 275, 2000
|
|
4UDJ
 
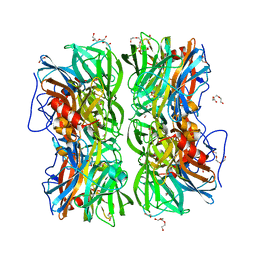 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.60 Angstrom in complex with beta-D-mannopyranose and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4TVD
 
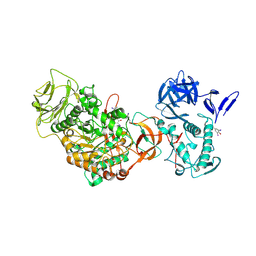 | | N-terminally truncated dextransucrase DSR-E from Leuconostoc mesenteroides NRRL B-1299 in complex with D-glucose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Dextransucrase, ... | | Authors: | Brison, Y, Remaud-Simeon, M, Mourey, L, Tranier, S. | | Deposit date: | 2014-06-26 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Carbohydrate Binding Ability of an alpha-(12) Branching Sucrase from Glycoside Hydrolase Family 70.
J.Biol.Chem., 291, 2016
|
|
4UDK
 
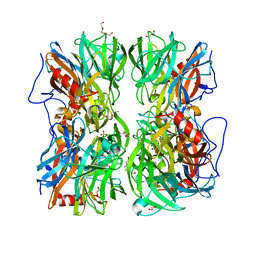 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.76 Angstrom from unknown human gut bacteria (Uhgb_MP) in complex with N-acetyl-D-glucosamine, beta-D-mannopyranose and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UDI
 
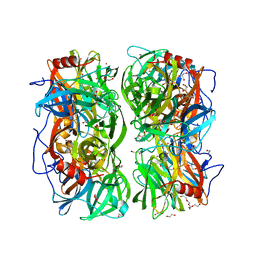 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.85 Angstrom from unknown human gut bacteria (Uhgb_MP) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UDG
 
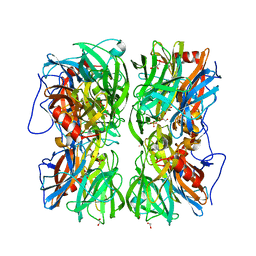 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.60 Angstrom in complex with N-acetylglucosamine and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
9CLB
 
 | | Crystal structure of Bak bound to the inhibitory aBAK | | Descriptor: | Bcl-2 homologous antagonist/killer, aBAK | | Authors: | Birkinshaw, R.W, Berger, S.A, Lee, E.F, Harris, T.J, Tran, S, Bera, A.K, Arguinchona, L, Kang, A, Sankaran, B, Kasapgil, S, Miller, M.S, Smyth, S, Uren, R, Kluck, R, Colman, P.M, Fairlie, W.D, Czabotar, P.E, Baker, D. | | Deposit date: | 2024-07-10 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Computational design of potent and selective inhibitors of BAK and BAX
To Be Published
|
|
4FLO
 
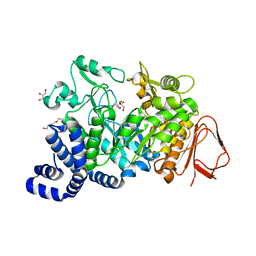 | | Crystal structure of Amylosucrase double mutant A289P-F290C from Neisseria polysaccharea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4FLS
 
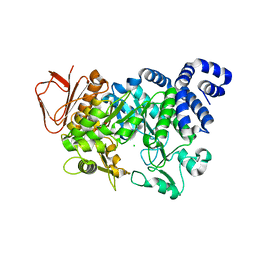 | | Crystal structure of Amylosucrase inactive double mutant F290K-E328Q from Neisseria polysaccharea in complex with sucrose. | | Descriptor: | Amylosucrase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4FLR
 
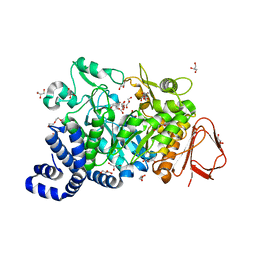 | | Crystal structure of Amylosucrase double mutant A289P-F290L from Neisseria polysaccharea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4FLQ
 
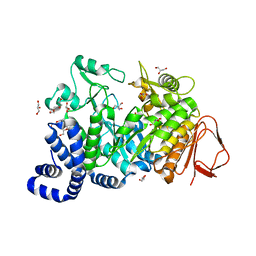 | | Crystal structure of Amylosucrase double mutant A289P-F290I from Neisseria polysaccharea. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
5N6V
 
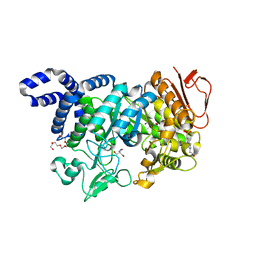 | | Crystal structure of Neisseria polysaccharea amylosucrase mutant derived from Neutral genetic Drift-based engineering | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amylosucrase, ... | | Authors: | Daude, D, Verges, A, Tranier, S. | | Deposit date: | 2017-02-16 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Neutral Genetic Drift-Based Engineering of a Sucrose-Utilizing Enzyme toward Glycodiversification.
Acs Catalysis, 2019
|
|
5N7J
 
 | |
5NHX
 
 | | Periplasmic domain of Outer Membrane Protein A from Klebsiella pneumoniae | | Descriptor: | CITRIC ACID, Outer membrane protein A | | Authors: | Demange, P, Tranier, S, Nars, G, Iordanov, I, Mourey, L, Saurel, O, Milon, A. | | Deposit date: | 2017-03-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and dynamics of the C-terminal domain of OmpA from Klebsiella pneumonia
To Be Published
|
|
8PWZ
 
 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadBD from Mycobacterium tuberculosis | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadB, UPF0336 protein Rv0504c | | Authors: | Rima, J, Grimoire, Y, Bories, P, Bardou, F, Quemard, A, Bon, C, Mourey, L, Tranier, S. | | Deposit date: | 2023-07-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.00196719 Å) | | Cite: | HadBD dehydratase from Mycobacterium tuberculosis fatty acid synthase type II: A singular structure for a unique function.
Protein Sci., 33, 2024
|
|
2Y7Y
 
 | | APLYSIA CALIFORNICA ACHBP IN APO STATE | | Descriptor: | SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, De Esch, I.J. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X-Ray Analysis.
J.Med.Chem., 52, 2009
|
|
3ZKR
 
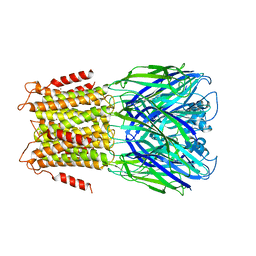 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromoform | | Descriptor: | CYS-LOOP LIGAND-GATED ION CHANNEL, TRIBROMOMETHANE | | Authors: | Spurny, R, Billen, B, Howard, R.J, Brams, M, Debaveye, S, Price, K.L, Weston, D.A, Strelkov, S.V, Tytgat, J, Bertrand, S, Bertrand, D, Lummis, S.C.R, Ulens, C. | | Deposit date: | 2013-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.649 Å) | | Cite: | Multisite Binding of a General Anesthetic to the Prokaryotic Pentameric Erwinia Chrysanthemi Ligand-Gated Ion Channel (Elic).
J.Biol.Chem., 288, 2013
|
|
3O0D
 
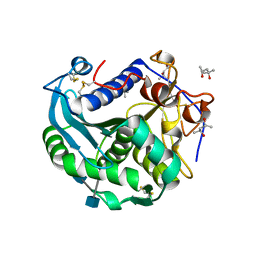 | | Crystal structure of Lip2 lipase from Yarrowia lipolytica at 1.7 A resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bordes, F, Tranier, S, Mourey, L, Marty, A. | | Deposit date: | 2010-07-19 | | Release date: | 2010-11-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring the conformational states and rearrangements of Yarrowia lipolytica Lipase.
Biophys.J., 99, 2010
|
|
4TTU
 
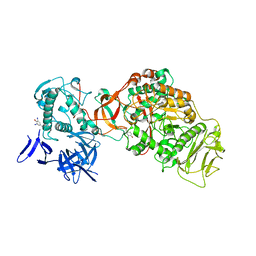 | | N-terminally truncated dextransucrase DSR-E from Leuconostoc mesenteroides NRRL B-1299 in complex with isomaltotriose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Dextransucrase, ... | | Authors: | Brison, Y, Remaud-Simeon, M, Mourey, L, Tranier, S. | | Deposit date: | 2014-06-23 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Insights into the Carbohydrate Binding Ability of an alpha-(12) Branching Sucrase from Glycoside Hydrolase Family 70.
J.Biol.Chem., 291, 2016
|
|
2QPS
 
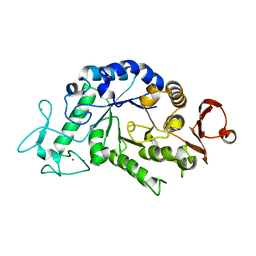 | | "Sugar tongs" mutant Y380A in complex with acarbose | | Descriptor: | Alpha-amylase type A isozyme, CALCIUM ION | | Authors: | Aghajari, N, Jensen, M.H, Tranier, S, Haser, R. | | Deposit date: | 2007-07-25 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 'pair of sugar tongs' site on the non-catalytic domain C of barley alpha-amylase participates in substrate binding and activity
Febs J., 274, 2007
|
|
4TVC
 
 | | N-terminally truncated dextransucrase DSR-E from Leuconostoc mesenteroides NRRL B-1299 in complex with gluco-oligosaccharides | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL, ... | | Authors: | Brison, Y, Remaud-Simeon, M, Mourey, L, Tranier, S. | | Deposit date: | 2014-06-26 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into the Carbohydrate Binding Ability of an alpha-(12) Branching Sucrase from Glycoside Hydrolase Family 70.
J.Biol.Chem., 291, 2016
|
|
2QPU
 
 | | Sugar tongs mutant S378P in complex with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 1,5-anhydro-4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S,6R)-2,3,4,6-tetrahydroxy-5-methylcyclohexyl]amino}-alpha-D-glucopyranosyl)-D-glucitol, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Aghajari, N, Jensen, M.H, Tranier, S, Haser, R. | | Deposit date: | 2007-07-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 'pair of sugar tongs' site on the non-catalytic domain C of barley alpha-amylase participates in substrate binding and activity
Febs J., 274, 2007
|
|
4N4B
 
 | | Crystal Structure of the alpha-L-arabinofuranosidase PaAbf62A from Podospora anserina | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
4N2R
 
 | | Crystal Structure of the alpha-L-arabinofuranosidase UmAbf62A from Ustilago maydis in complex with L-arabinofuranose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, alpha-L-arabinofuranose, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-06 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
