8D02
 
 | | Crystal Structure of Bacillus anthracis BxpB | | Descriptor: | BxpB, CALCIUM ION | | Authors: | Chattopadhyay, D, Turnbough, C.L. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and induced stability of trimeric BxpB: implications for the assembly of BxpB-BclA complexes in the exosporium of Bacillus anthracis.
Mbio, 14, 2023
|
|
2LKW
 
 | |
5UI1
 
 | | Crystal Structure of Human Protein Phosphatase 5C (PP5C) in complex with a triazole inhibitor | | Descriptor: | 5-phenyl-1H-1,2,3-triazole-4-carboxylic acid, MANGANESE (II) ION, Serine/threonine-protein phosphatase 5 | | Authors: | Chattopadhyay, D, Swingle, M.R, Salter, E.A, Banerjee, S, Honkanen, R.E. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure Human PP5C in Complex with an Inhibitor
To Be Published
|
|
4ZX2
 
 | | Co-crystal structures of PP5 in complex with 5-methyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid | | Descriptor: | (1S,2R,3S,4R,5S)-5-methyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Chattopadhyay, D, Swingle, M.R, Salter, E.A, Wierzbicki, A, Honkanen, R.E. | | Deposit date: | 2015-05-19 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal structures and mutagenesis of PPP-family ser/thr protein phosphatases elucidate the selectivity of cantharidin and novel norcantharidin-based inhibitors of PP5C.
Biochem. Pharmacol., 109, 2016
|
|
4ZVZ
 
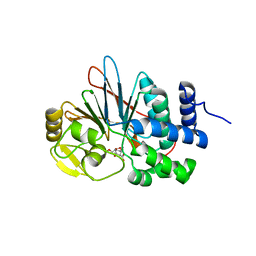 | | Co-crystal structures of PP5 in complex with 5-methyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid | | Descriptor: | (1R,2S,3R,4S,5S)-5-(propoxymethyl)-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Chattopadhyay, D, Swingle, M.R, Salter, E.A, Wierzbicki, A, Honkanen, R.E. | | Deposit date: | 2015-05-18 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and mutagenesis of PPP-family ser/thr protein phosphatases elucidate the selectivity of cantharidin and novel norcantharidin-based inhibitors of PP5C.
Biochem. Pharmacol., 109, 2016
|
|
1D5C
 
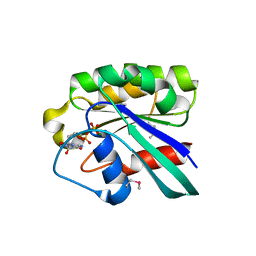 | | CRYSTAL STRUCTURE OF PLASMODIUM FALCIPARUM RAB6 COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAB6 GTPASE | | Authors: | Chattopadhyay, D, Langsley, G, Carson, M, Recacha, R, DeLucas, L, Smith, C. | | Deposit date: | 1999-10-06 | | Release date: | 2000-08-30 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the nucleotide-binding domain of Plasmodium falciparum rab6 in the GDP-bound form.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3CLV
 
 | | Crystal Structure of Rab5a from plasmodium falciparum, PFB0500c | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Rab5 protein, ... | | Authors: | Chattopadhyay, D, Wernimont, A.K, Langsley, G, Lew, J, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Sukumar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-20 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Rab5a from plasmodium falciparum, PFB0500c
To be Published
|
|
1LTK
 
 | |
5MQG
 
 | |
5MPZ
 
 | |
5MGK
 
 | |
5MGG
 
 | |
5MGJ
 
 | |
5MGL
 
 | |
5MQE
 
 | |
5MGE
 
 | |
5MQK
 
 | |
5MGF
 
 | |
5MGM
 
 | |
3NT7
 
 | |
7UW2
 
 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with UAB116 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-8-{3-[(2S)-butan-2-yl]-2-(3-methylbutyl)cyclohex-2-en-1-ylidene}-3,7-dimethylocta-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chattopadhyay, D, Yang, Z, Atigadda, V. | | Deposit date: | 2022-05-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conformationally Defined Rexinoids for the Prevention of Inflammation and Nonmelanoma Skin Cancers.
J.Med.Chem., 65, 2022
|
|
7UW4
 
 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with UAB113 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-[2-(3-methylbutyl)-3-propylcyclohex-2-en-1-ylidene]octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chattopadhyay, D, Yang, Z, Atigadda, V. | | Deposit date: | 2022-05-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformationally Defined Rexinoids for the Prevention of Inflammation and Nonmelanoma Skin Cancers.
J.Med.Chem., 65, 2022
|
|
4ND5
 
 | |
4ND2
 
 | |
4ND1
 
 | |
