5GTT
 
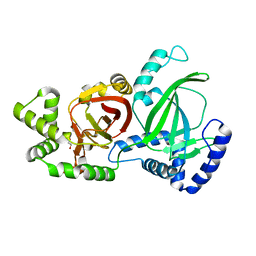 | | Crystal structure of C. perfringens iota-like enterotoxin CPILE-a | | Descriptor: | 1,2-ETHANEDIOL, Binary enterotoxin of Clostridium perfringens component a | | Authors: | Toniti, W, Yoshida, T, Tsurumura, T, Irikura, D, Tsuge, H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal structure and structure-based mutagenesis of actin-specific ADP-ribosylating toxin CPILE-a as novel enterotoxin
PLoS ONE, 12, 2017
|
|
5WTZ
 
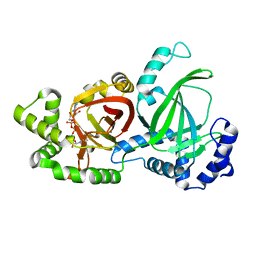 | | Crystal structure of C. perfringens iota-like enterotoxin CPILE-a with NAD+ | | Descriptor: | Binary enterotoxin of Clostridium perfringens component a, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Toniti, W, Yoshida, T, Tsurumura, T, Irikura, D, Tsuge, H. | | Deposit date: | 2016-12-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal structure and structure-based mutagenesis of actin-specific ADP-ribosylating toxin CPILE-a as novel enterotoxin
PLoS ONE, 12, 2017
|
|
5WU0
 
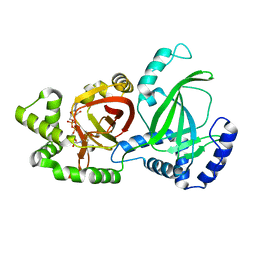 | | Crystal structure of C. perfringens iota-like enterotoxin CPILE-a with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Binary enterotoxin of Clostridium perfringens component a | | Authors: | Toniti, W, Yoshida, T, Tsurumura, T, Irikura, D, Tsuge, H. | | Deposit date: | 2016-12-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Crystal structure and structure-based mutagenesis of actin-specific ADP-ribosylating toxin CPILE-a as novel enterotoxin
PLoS ONE, 12, 2017
|
|
1GL2
 
 | | Crystal structure of an endosomal SNARE core complex | | Descriptor: | ENDOBREVIN, SYNTAXIN 7, SYNTAXIN 8, ... | | Authors: | Antonin, W, Becker, S, Jahn, R, Schneider, T.R. | | Deposit date: | 2001-08-22 | | Release date: | 2002-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Endosomal Snare Complex Reveals Common Structural Principles of All Snares.
Nat.Struct.Biol., 9, 2001
|
|
5FKI
 
 | | Pseudorabies virus (PrV) nuclear egress complex proteins fitted as a hexameric lattice into a sub-tomogram average derived from focused- ion beam milled lamellae electron cryo-microscopic data | | Descriptor: | CHLORIDE ION, UL31, UL34 protein, ... | | Authors: | Hagen, C, Dent, K.C, Zeev Ben Mordehai, T, Vasishtan, D, Antonin, W, Mettenleiter, T.C, Gruenewald, K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights Into Inner Nuclear Membrane Remodelling
Cell Rep., 13, 2015
|
|
5A9Q
 
 | | Human nuclear pore complex | | Descriptor: | NUCLEAR PORE COMPLEX PROTEIN NUP107, NUCLEAR PORE COMPLEX PROTEIN NUP133, NUCLEAR PORE COMPLEX PROTEIN NUP155, ... | | Authors: | von Appen, A, Kosinski, J, Sparks, L, Ori, A, DiGuilio, A, Vollmer, B, Mackmull, M, Banterle, N, Parca, L, Kastritis, P, Buczak, K, Mosalaganti, S, Hagen, W, Andres-Pons, A, Lemke, E.A, Bork, P, Antonin, W, Glavy, J.S, Bui, K.H, Beck, M. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | In Situ Structural Analysis of the Human Nuclear Pore Complex
Nature, 526, 2015
|
|
9B0A
 
 | |
8FCY
 
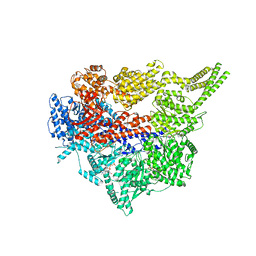 | | Engineered human dynein motor domain in microtubule-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase | | Authors: | Ton, W, Wang, Y, Chai, P. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FDT
 
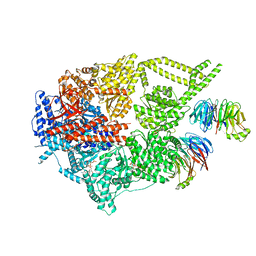 | | Engineered human dynein motor domain in the microtubule-unbound state with LIS1 complex in the buffer containing ATP-Vi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase, ... | | Authors: | Ton, W, Wang, Y, Chai, P. | | Deposit date: | 2022-12-04 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FD6
 
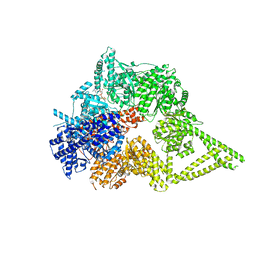 | | Engineered human dynein motor domain in the microtubule-unbound state in the buffer containing ATP-Vi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase, ... | | Authors: | Ton, W, Wang, Y, Chai, P. | | Deposit date: | 2022-12-02 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FDU
 
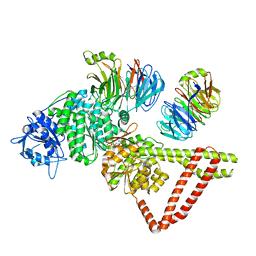 | | Engineered human dynein motor domain in the microtubule-unbound state with LIS1 complex in the buffer containing ATP-Vi (local refined on AAA3-AAA5 and LIS1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase, Platelet-activating factor acetylhydrolase IB subunit beta,Platelet-activating factor acetylhydrolase IB subunit beta,human LIS1 protein with a SNAP tag | | Authors: | Ton, W, Wang, Y, Chai, P. | | Deposit date: | 2022-12-04 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1AV8
 
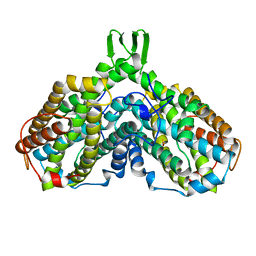 | | RIBONUCLEOTIDE REDUCTASE R2 SUBUNIT FROM E. COLI | | Descriptor: | MU-OXO-DIIRON, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Han, S, Arvai, A, Tainer, J.A. | | Deposit date: | 1997-09-30 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of Y122F R2 of Escherichia coli ribonucleotide reductase by time-resolved physical biochemical methods and X-ray crystallography.
Biochemistry, 37, 1998
|
|
7RML
 
 | | Neisseria meningitidis Methylenetetrahydrofolate reductase in complex with FAD | | Descriptor: | 5,10-methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pederick, J.L, Wegener, K.L, Salaemae, W, Bruning, J.B. | | Deposit date: | 2021-07-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural characterization of meningococcal methylenetetrahydrofolate reductase.
Protein Sci., 32, 2023
|
|
2AV8
 
 | | Y122F MUTANT OF RIBONUCLEOTIDE REDUCTASE FROM ESCHERICHIA COLI | | Descriptor: | FE (II) ION, MU-OXO-DIIRON, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Han, S, Arvai, A, Tainer, J.A. | | Deposit date: | 1997-09-30 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Characterization of Y122F R2 of Escherichia coli ribonucleotide reductase by time-resolved physical biochemical methods and X-ray crystallography.
Biochemistry, 37, 1998
|
|
2DHB
 
 | |
8Q2D
 
 | |
8Q2C
 
 | |
2WON
 
 | | Crystal Structure of UK-453061 bound to HIV-1 Reverse Transcriptase (wild-type). | | Descriptor: | 5-{[3,5-diethyl-1-(2-hydroxyethyl)-1H-pyrazol-4-yl]oxy}benzene-1,3-dicarbonitrile, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Phillips, C, Irving, S.L, Knoechel, T, Ringrose, H. | | Deposit date: | 2009-07-27 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lersivirine: A Non-Nucleoside Reverse Transcriptase Inhibitor with Activity Against Drug- Resistant Human Immunodeficiency Virus-1.
Antimicrob.Agents Chemother., 54, 2010
|
|
5E8C
 
 | | pseudorabies virus nuclear egress complex, pUL31, pUL34 | | Descriptor: | CHLORIDE ION, UL31, UL34 protein, ... | | Authors: | Zeev-Ben-Mordehai, T, Cheleski, J, Whittle, C, El Omari, K, Harlos, K, Hagen, C, Klupp, B, Mettenleiter, T.C, Gruenewald, K. | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights into Inner Nuclear Membrane Remodeling.
Cell Rep, 13, 2015
|
|
5J0N
 
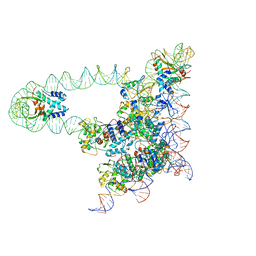 | | Lambda excision HJ intermediate | | Descriptor: | Excisionase, Integrase, Integration host factor subunit alpha, ... | | Authors: | Van Duyne, G, Grigorieff, N, Landy, A. | | Deposit date: | 2016-03-28 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structure of a Holliday junction complex reveals mechanisms governing a highly regulated DNA transaction.
Elife, 5, 2016
|
|
3KW9
 
 | |
3KWZ
 
 | |
3KX1
 
 | |
3OVX
 
 | |
3OVZ
 
 | | Cathepsin K in complex with a covalent inhibitor with a ketoamide warhead | | Descriptor: | Cathepsin K, N-[(1S)-3-amino-1-ethyl-2,3-dioxopropyl]-2-chloro-4-(pyridin-2-ylmethoxy)-3-(trifluoromethyl)benzamide, SULFATE ION | | Authors: | Fradera, X, van Zeeland, M, Uitdehaag, J.C.M. | | Deposit date: | 2010-09-17 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Trifluoromethylphenyl as P2 for ketoamide-based cathepsin S inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
