7ATY
 
 | | Solution NMR structure of the SH3 domain of human Caskin1 | | Descriptor: | Caskin-1 | | Authors: | Toke, O, Koprivanacz, K, Radnai, L, Mero, B, Juhasz, T, Liliom, K, Buday, L. | | Deposit date: | 2020-11-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the SH3 Domain of Human Caskin1 Validates the Lack of a Typical Peptide Binding Groove and Supports a Role in Lipid Mediator Binding.
Cells, 10, 2021
|
|
2MHW
 
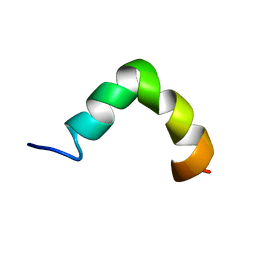 | | The solution NMR structure of maximin-4 in SDS micelles | | Descriptor: | Antimicrobial peptide | | Authors: | Toke, O, Banoczi, Z, Kiraly, P, Heinzmann, R, Burck, J, Ulrich, A.S, Hudecz, F. | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A kinked antimicrobial peptide from Bombina maxima. I. Three-dimensional structure determined by NMR in membrane-mimicking environments.
Eur.Biophys.J., 40, 2011
|
|
2MM3
 
 | | Solution NMR structure of the ternary complex of human ileal bile acid-binding protein with glycocholate and glycochenodeoxycholate | | Descriptor: | GLYCOCHENODEOXYCHOLIC ACID, GLYCOCHOLIC ACID, Gastrotropin | | Authors: | Horvath, G, Egyed, O, Bencsura, A, Simon, A, Tochtrop, G.P, DeKoster, G.T, Covey, D.F, Cistola, D.P, Toke, O. | | Deposit date: | 2014-03-07 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of ligand binding in the ternary complex of human ileal bile acid binding protein with glycocholate and glycochenodeoxycholate obtained from solution NMR
FEBS J., 283, 2016
|
|
6ZQS
 
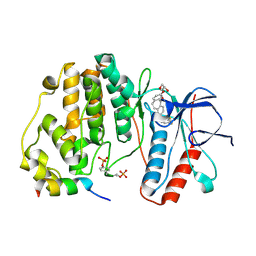 | | Crystal structure of double-phosphorylated p38alpha with ATF2(83-102) | | Descriptor: | 2-[(2,4-difluorophenyl)amino]-7-{[(2R)-2,3-dihydroxypropyl]oxy}-10,11-dihydro-5H-dibenzo[a,d][7]annulen-5-one, Cyclic AMP-dependent transcription factor ATF-2, Mitogen-activated protein kinase 14 | | Authors: | Kirsch, K, Sok, P, Poti, A.L, Remenyi, A. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Co-regulation of the transcription controlling ATF2 phosphoswitch by JNK and p38.
Nat Commun, 11, 2020
|
|
6ZR5
 
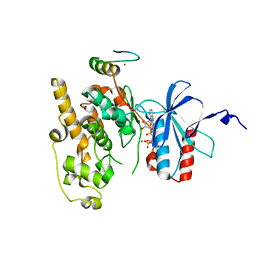 | | Crystal structure of JNK1 in complex with ATF2(19-58) | | Descriptor: | Cyclic AMP-dependent transcription factor ATF-2, MAGNESIUM ION, Mitogen-activated protein kinase 8, ... | | Authors: | Kirsch, K, Zeke, A, Remenyi, A. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Co-regulation of the transcription controlling ATF2 phosphoswitch by JNK and p38.
Nat Commun, 11, 2020
|
|
5NP3
 
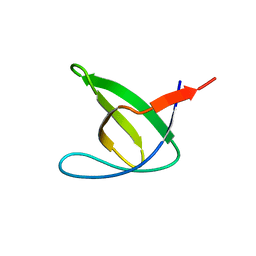 | | Abl2 SH3 | | Descriptor: | Abelson tyrosine-protein kinase 2 | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
5NP5
 
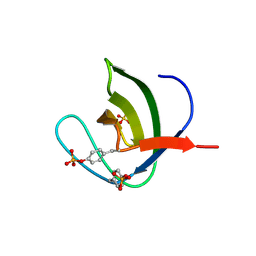 | | Abl2 SH3 pTyr116/161 | | Descriptor: | Abelson tyrosine-protein kinase 2, SULFATE ION | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
5NP2
 
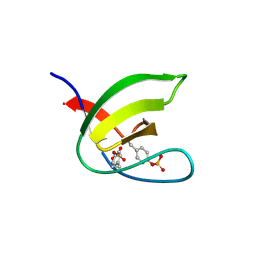 | | Abl1 SH3 pTyr89/134 | | Descriptor: | Tyrosine-protein kinase ABL1 | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
