5M2D
 
 | |
5M0K
 
 | |
5M6G
 
 | | Crystal structure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea | | Descriptor: | Beta-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.829 Å) | | Cite: | Crystal structure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea
To Be Published
|
|
5MRJ
 
 | | Crystal structure of Endo-1,4-beta-xylanase-like protein from Acremonium chrysogenum | | Descriptor: | Beta-xylanase, SULFATE ION | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-12-23 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Endo-1,4-beta-xylanase-like protein from Acremonium chrysogenum
To Be Published
|
|
5MRS
 
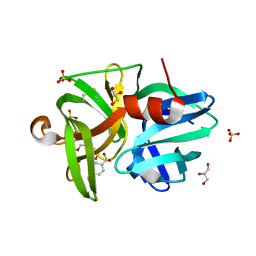 | | Crystal structure of L1 protease Lysobacter sp. XL1 in complex with AEBSF | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of L1 protease Lysobacter sp. XL1 in complex with AEBSF
To Be Published
|
|
5MRR
 
 | | Crystal structure of L1 protease of Lysobacter sp. XL1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of L1 protease of Lysobacter sp. XL1
To Be Published
|
|
5MRT
 
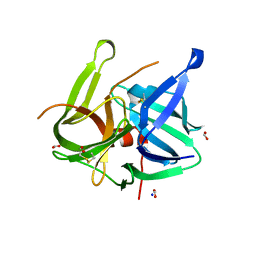 | | Crystal structure of L5 protease Lysobacter sp. XL1 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of L5 protease Lysobacter sp. XL1
To Be Published
|
|
4REO
 
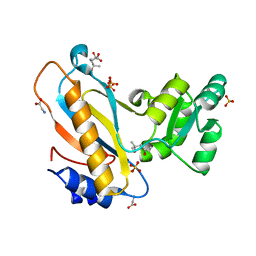 | | Mutant ribosomal protein l1 from thermus thermophilus with threonine 217 replaced by valine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L1, GLYCINE, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Tishchenko, S.V, Nikonov, S.V. | | Deposit date: | 2014-09-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QGB
 
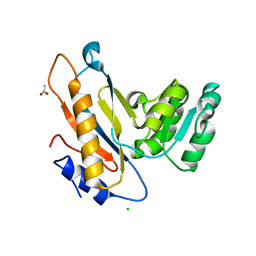 | | Crystal structure of mutant ribosomal protein G219V TthL1 | | Descriptor: | 50S ribosomal protein L1, ACETATE ION, CHLORIDE ION | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3U4M
 
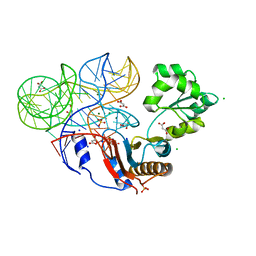 | | Crystal structure of ribosomal protein tthl1 in complex with 80nt 23s rna from thermus thermophilus | | Descriptor: | 50S ribosomal protein L1, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, NIkonov, S.V. | | Deposit date: | 2011-10-10 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structure of the isolated ribosomal L1 stalk.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4QG3
 
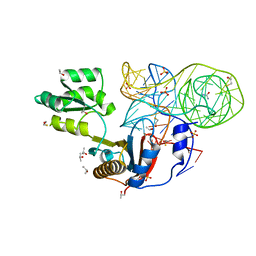 | | Crystal structure of mutant ribosomal protein G219V TthL1 in complex with 80nt 23S RNA from Thermus thermophilus | | Descriptor: | 50S ribosomal protein L1, BETA-MERCAPTOETHANOL, ISOPROPYL ALCOHOL, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QVI
 
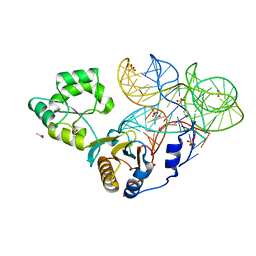 | | Crystal structure of mutant ribosomal protein M218L TthL1 in complex with 80nt 23S RNA from Thermus thermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 50S ribosomal protein L1, ACETATE ION, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, NIkonov, S.V. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3TG8
 
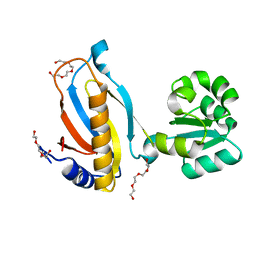 | | Mutant ribosomal protein L1 lacking ala158 from thermus thermophilus | | Descriptor: | 50S ribosomal protein L1, CHLORIDE ION, TETRAETHYLENE GLYCOL | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2011-08-17 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of interdomain mobility in ribosomal L1 proteins.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1AN7
 
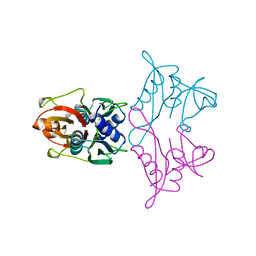 | |
5DY9
 
 | | Y68T Hfq from Methanococcus jannaschii in complex with AMP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nikulin, A.D, Mikhailina, A.O, Lekontseva, N.V, Balobanov, V.A, Nikonova, E.Y, Tishchenko, S.V. | | Deposit date: | 2015-09-24 | | Release date: | 2016-09-28 | | Last modified: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
4X9C
 
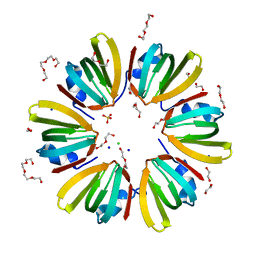 | | 1.4A crystal structure of Hfq from Methanococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Tishchenko, S.V, Nikonova, S.V, Murina, V.N, Mihailina, A.O, Lekontseva, N.V. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
4X9D
 
 | | High-resolution structure of Hfq from Methanococcus jannaschii in complex with UMP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Tishchenko, S.V, Nikonova, E.Y, Murina, V.N, Mihailina, A.O, Lekontseva, N.V. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
8AF1
 
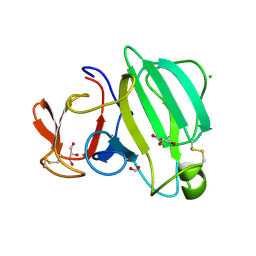 | | Beta-Lytic Protease from Lysobacter capsici | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kudryakova, I.V, Afoshin, A.S, Vasilyeva, N.V. | | Deposit date: | 2022-07-15 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural and Functional Characterization of beta-lytic Protease from Lysobacter capsici VKM B-2533 T.
Int J Mol Sci, 23, 2022
|
|
8B7W
 
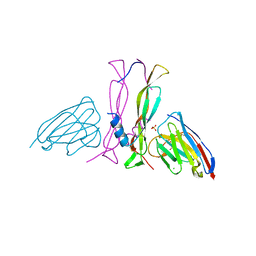 | | Complex IL-17A/anti-IL-17A-76 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-17A, ... | | Authors: | Kostareva, O.S, Svoeglazova, A, Kolyadenko, I.A, Dzhus, U.F, Tishchenko, S.V, Gabdulkhakov, A.G. | | Deposit date: | 2022-10-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Two Epitope Regions Revealed in the Complex of IL-17A and Anti-IL-17A V H H Domain.
Int J Mol Sci, 23, 2022
|
|
8P9U
 
 | | Crystal Structure of Two-Domain Laccase mutant M199A/D268N from Streptomyces griseoflavus | | Descriptor: | CALCIUM ION, COPPER (II) ION, OXYGEN MOLECULE, ... | | Authors: | Kolyadenko, I.A, Tishchenko, S.V, Gabdulkhakov, A.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight into the Amino Acid Environment of the Two-Domain Laccase's Trinuclear Copper Cluster.
Int J Mol Sci, 24, 2023
|
|
6FC7
 
 | |
6FDJ
 
 | |
6ZIP
 
 | | Crystal Structure of Two-Domain Laccase mutant R240A from Streptomyces griseoflavus | | Descriptor: | COPPER (II) ION, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The role of positive charged residue in the proton-transfer mechanism of two-domain laccase from Streptomyces griseoflavus Ac-993.
J.Biomol.Struct.Dyn., 40, 2022
|
|
6ZIJ
 
 | | Crystal Structure of Two-Domain Laccase mutant R240H from Streptomyces griseoflavus | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The role of positive charged residue in the proton-transfer mechanism of two-domain laccase from Streptomyces griseoflavus Ac-993.
J.Biomol.Struct.Dyn., 40, 2022
|
|
8P9V
 
 | | Crystal Structure of Two-Domain Laccase mutant M199G/R240H/D268N from Streptomyces griseoflavus | | Descriptor: | COPPER (II) ION, OXYGEN MOLECULE, PEROXIDE ION, ... | | Authors: | Kolyadenko, I.A, Tishchenko, S.V, Gabdulkhakov, A.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into the Amino Acid Environment of the Two-Domain Laccase's Trinuclear Copper Cluster.
Int J Mol Sci, 24, 2023
|
|
