2BVD
 
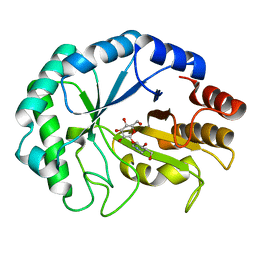 | | HOW FAMILY 26 GLYCOSIDE HYDROLASES ORCHESTRATE CATALYSIS ON DIFFERENT POLYSACCHARIDES. STRUCTURE AND ACTIVITY OF A CLOSTRIDIUM THERMOCELLUM LICHENASE, CtLIC26A | | Descriptor: | (3R,4R,5R)-4-hydroxy-5-(hydroxymethyl)piperidin-3-yl beta-D-glucopyranoside, ENDOGLUCANASE H | | Authors: | Taylor, E.J, Goyal, A, Guerreiro, C.I.P.D, Prates, J.A.M, Money, V.A, Ferry, N, Morland, C, Planas, A, Macdonald, J.A, Stick, R.V, Gilbert, H.J, Fontes, C.M.G.A, Davies, G.J. | | Deposit date: | 2005-06-27 | | Release date: | 2005-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How Family 26 Glycoside Hydrolases Orchestrate Catalysis on Different Polysaccharides: Structure and Activity of a Clostridium Thermocellum Lichenase, Ctlic26A.
J.Biol.Chem., 280, 2005
|
|
2BV9
 
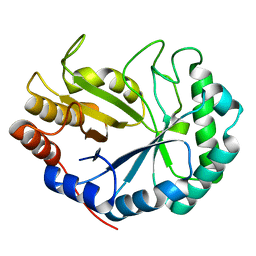 | | HOW FAMILY 26 GLYCOSIDE HYDROLASES ORCHESTRATE CATALYSIS ON DIFFERENT POLYSACCHARIDES. STRUCTURE AND ACTIVITY OF A CLOSTRIDIUM THERMOCELLUM LICHENASE, CtLIC26A | | Descriptor: | ENDOGLUCANASE H | | Authors: | Taylor, E.J, Goyal, A, Guerreiro, C.I.P.D, Prates, J.A.M, Money, V.A, Ferry, N, Morland, C, Planas, A, Macdonald, J.A, Stick, R.V, Gilbert, H.J, Fontes, C.M.G.A, Davies, G.J. | | Deposit date: | 2005-06-23 | | Release date: | 2005-06-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How Family 26 Glycoside Hydrolases Orchestrate Catalysis on Different Polysaccharides: Structure and Activity of a Clostridium Thermocellum Lichenase, Ctlic26A.
J.Biol.Chem., 280, 2005
|
|
2CC0
 
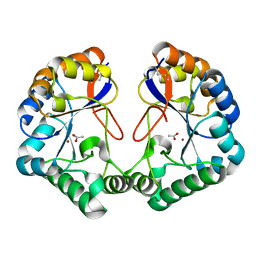 | | Family 4 carbohydrate esterase from Streptomyces lividans in complex with acetate | | Descriptor: | ACETATE ION, ACETYL-XYLAN ESTERASE, ZINC ION | | Authors: | Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Vincent, F, Brzozowski, A.M, Dupont, C, Shareck, F, Centeno, M.S.J, Prates, J.A.M, Puchart, V, Ferreira, L.M.A, Fontes, C.M.G.A, Biely, P, Davies, G.J. | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Activity of Two Metal-Ion Dependent Acetyl Xylan Esterases Involved in Plant Cell Wall Degradation Reveals a Close Similarity to Peptidoglycan Deacetylases
J.Biol.Chem., 281, 2006
|
|
2C8N
 
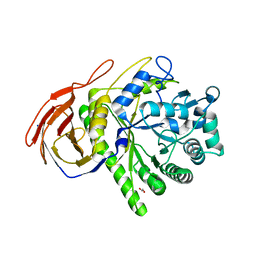 | | The Structure of a family 51 arabinofuranosidase, Araf51, from Clostridium thermocellum in complex with 1,3-linked arabinoside of xylobiose. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, alpha-L-arabinofuranose-(1-3)-alpha-D-xylopyranose | | Authors: | Taylor, E.J, Smith, N.L, Turkenburg, J.P, D'Souza, S, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-12-06 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight Into the Ligand Specificity of a Thermostable Family 51 Arabinofuranosidase, Araf51, from Clostridium Thermocellum.
Biochem.J., 395, 2006
|
|
2C7F
 
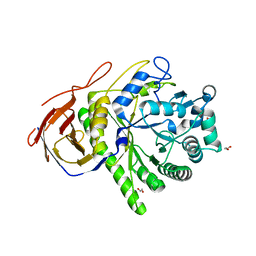 | | The Structure of a family 51 arabinofuranosidase, Araf51, from Clostridium thermocellum in complex with 1,5-alpha-L-Arabinotriose. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, ... | | Authors: | Taylor, E.J, Smith, N.L, Turkenburg, J.P, D'Souza, S, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-11-23 | | Release date: | 2005-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight Into the Ligand Specificity of a Thermostable Family 51 Arabinofuranosidase, Araf51, from Clostridium Thermocellum.
Biochem.J., 395, 2006
|
|
2C79
 
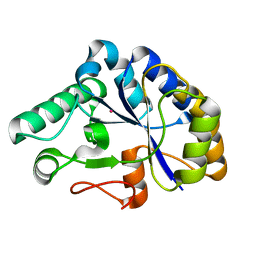 | | The structure of a family 4 acetyl xylan esterase from Clostridium thermocellum in complex with a colbalt ion. | | Descriptor: | COBALT (II) ION, GLYCOSIDE HYDROLASE, FAMILY 11:CLOSTRIDIUM CELLULOSOME ENZYME, ... | | Authors: | Taylor, E.J, Turkenburg, P.J, Vincent, F, Brzozowski, A.M, Gloster, T.M, Dupont, C, Shareck, F, Centeno, M.S.J, Prates, J.A.M, Ferreira, L.M.A, Fontes, C.M.G.A, Biely, P, Davies, G.J. | | Deposit date: | 2005-11-18 | | Release date: | 2006-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Activity of Two Metal-Ion Dependent Acetyl Xylan Esterases Involved in Plant Cell Wall Degradation Reveals a Close Similarity to Peptidoglycan Deacetylases.
J.Biol.Chem., 281, 2006
|
|
2C71
 
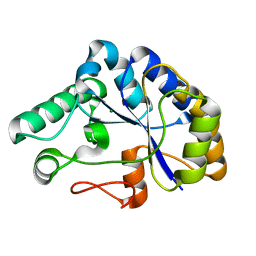 | | The structure of a family 4 acetyl xylan esterase from Clostridium thermocellum in complex with a magnesium ion. | | Descriptor: | GLYCOSIDE HYDROLASE, FAMILY 11:CLOSTRIDIUM CELLULOSOME ENZYME, DOCKERIN TYPE I:POLYSACCHARIDE, ... | | Authors: | Taylor, E.J, Turkenburg, P.J, Vincent, F, Brzozowski, A.M, Gloster, T.M, Dupont, C, Shareck, F, Centeno, M.S.J, Prates, J.A.M, Ferreira, L.M.A, Fontes, C.M.G.A, Biely, P, Davies, G.J. | | Deposit date: | 2005-11-17 | | Release date: | 2006-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure and Activity of Two Metal-Ion Dependent Acetyl Xylan Esterases Involved in Plant Cell-Wall Degradation Reveals a Close Similarity to Peptidoglycan Deacetylases.
J.Biol.Chem., 281, 2006
|
|
6T5S
 
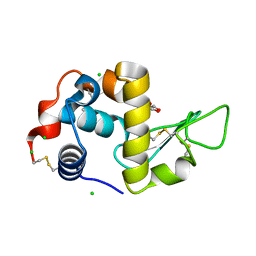 | | Apo form of C-type lysozyme from the upper gastrointestinal tract of Opisthocomus hoatzin | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C | | Authors: | Taylor, E.J, Skjot, M, Skov, L.K, Klausen, M, De Maria, L, Gippert, G.P, Turkenburg, J.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-10-17 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-Type Lysozyme from the upper Gastrointestinal Tract of Opisthocomus hoatzin, the Stinkbird.
Int J Mol Sci, 20, 2019
|
|
6T6C
 
 | | Complex with chitin oligomer of C-type lysozyme from the upper gastrointestinal tract of Opisthocomus hoatzin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C | | Authors: | Taylor, E.J, Skjot, M, Skov, L.K, Klausen, M, De Maria, L, Gippert, G.P, Turkenburg, J.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-10-18 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The C-Type Lysozyme from the upper Gastrointestinal Tract of Opisthocomus hoatzin, the Stinkbird.
Int J Mol Sci, 20, 2019
|
|
2C3F
 
 | | The structure of a group A streptococcal phage-encoded tail-fibre showing hyaluronan lyase activity. | | Descriptor: | HYALURONIDASE, PHAGE ASSOCIATED, SODIUM ION | | Authors: | Taylor, E.J, Smith, N.L, Linsay, A.-M, Charnock, S.J, Turkenburg, J.P, Dodson, E.J, Davies, G.J, Black, G.W. | | Deposit date: | 2005-10-06 | | Release date: | 2005-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of a Group a Streptococcal Phage-Encoded Virulence Factor Reveals Catalytically Active Triple-Stranded Beta-Helix
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1W2P
 
 | | The 3-dimensional structure of a xylanase (Xyn10A) from Cellvibrio japonicus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDO-1,4-BETA-XYLANASE A PRECURSOR | | Authors: | Taylor, E.J, Vincent, F, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2004-07-07 | | Release date: | 2004-09-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Use of Forced Protein Evolution to Investigate and Improve Stability of Family 10 Xylanases: The Production of Ca2+-Independent Stable Xylanases
J.Biol.Chem., 279, 2004
|
|
2WLY
 
 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitotrio-thiazoline. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-29 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chitinase inhibition by chitobiose and chitotriose thiazolines.
Angew. Chem. Int. Ed. Engl., 49, 2010
|
|
2WM0
 
 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitobio- thiazoline thioamide. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-29 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chitinase Inhibition by Chitobiose and Chitotriose Thiazolines.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2WK2
 
 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitotrio-thiazoline dithioamide. | | Descriptor: | 2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose-(1-4)-2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, CHITINASE A, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-04 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chitinase Inhibition by Chitobiose and Chitotriose Thiazolines.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2WLZ
 
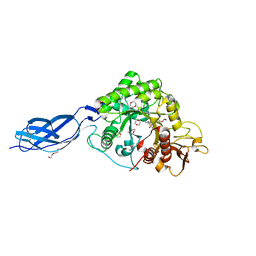 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitobio- thiazoline. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-29 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Chitinase Inhibition by Chitobiose and Chitotriose Thiazolines.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
8AM3
 
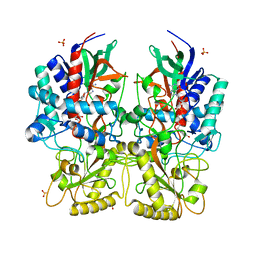 | |
8AM8
 
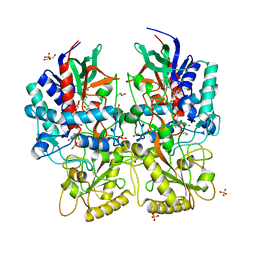 | |
8AM6
 
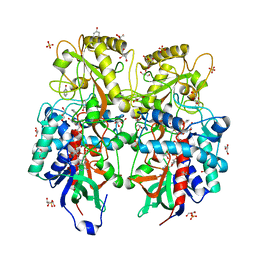 | |
1GYH
 
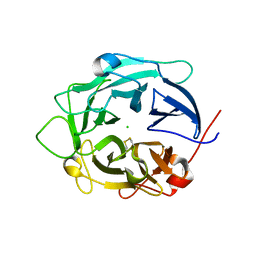 | | Structure of D158A Cellvibrio cellulosa alpha-L-arabinanase mutant | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A, CHLORIDE ION | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Cellovibrio Cellulosa Alpha-L-Arabinanase 43A Has a Novel Five-Blade Beta-Propeller Fold
Nat.Struct.Biol., 9, 2002
|
|
1GYD
 
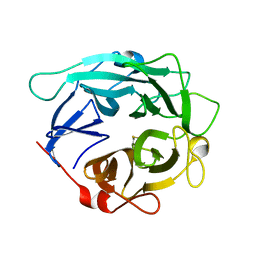 | | Structure of Cellvibrio cellulosa alpha-L-arabinanase | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade beta-propeller fold.
Nat. Struct. Biol., 9, 2002
|
|
1GYE
 
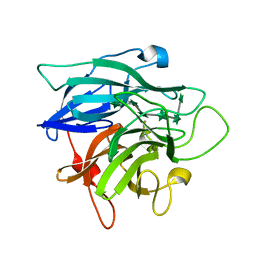 | | Structure of Cellvibrio cellulosa alpha-L-arabinanase complexed with Arabinohexaose | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A, CHLORIDE ION, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade beta-propeller fold.
Nat. Struct. Biol., 9, 2002
|
|
4B3L
 
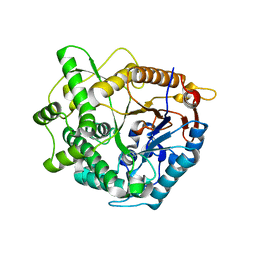 | | Family 1 6-phospho-beta-D glycosidase from Streptococcus pyogenes | | Descriptor: | BETA-GLUCOSIDASE | | Authors: | Stepper, J, Dabin, J, Ekloef, J.M, Thongpoo, P, Kongsaeree, P.T, Taylor, E.J, Turkenburg, J.P, Brumer, H, Davies, G.J. | | Deposit date: | 2012-07-24 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure and Activity of the Streptococcus Pyogenes Family Gh1 6-Phospho Beta-Glycosidase Spy1599
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4B3K
 
 | | Family 1 6-phospho-beta-D glycosidase from Streptococcus pyogenes | | Descriptor: | BETA-GLUCOSIDASE | | Authors: | Stepper, J, Dabin, J, Ekloef, J.M, Thongpoo, P, Kongsaeree, P.T, Taylor, E.J, Turkenburg, J.P, Brumer, H, Davies, G.J. | | Deposit date: | 2012-07-24 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Activity of the Streptococcus Pyogenes Family Gh1 6-Phospho Beta-Glycosidase Spy1599
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1WCU
 
 | | CBM29_1, A Family 29 Carbohydrate Binding Module from Piromyces equi | | Descriptor: | GLYCEROL, NON-CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davis, G.J, Gilbert, H.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
2CN2
 
 | | Crystal Structures of Clostridium thermocellum Xyloglucanase | | Descriptor: | BETA-1,4-XYLOGLUCAN HYDROLASE, CADMIUM ION | | Authors: | Martinez-Fleites, C, Taylor, E.J, Guerreiro, C.I, Prates, J.A.M, Ferreira, L.M.A, Fontes, C.M.G.A, Baumann, M.J, Brumer, H, Davies, G.J. | | Deposit date: | 2006-05-17 | | Release date: | 2006-05-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Clostridium Thermocellum Xyloglucanase, Xgh74A, Reveal the Structural Basis for Xyloglucan Recognition and Degradation.
J.Biol.Chem., 281, 2006
|
|
