6IWH
 
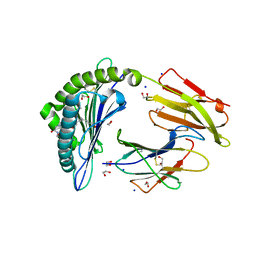 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with C14-GGGI lipopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, C14-GGGI lipopeptide, ... | | Authors: | Yamamoto, Y, Morita, D, Sugita, M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification and Structure of an MHC Class I-Encoded Protein with the Potential to PresentN-Myristoylated 4-mer Peptides to T Cells.
J Immunol., 202, 2019
|
|
6IWG
 
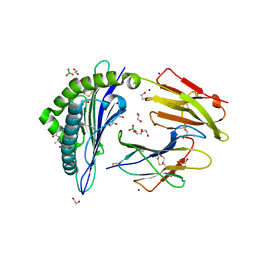 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with N-myristoylated 4-mer lipopeptide derived from SIV nef protein | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, Beta-2-microglobulin, ... | | Authors: | Yamamoto, Y, Morita, D, Sugita, M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Structure of an MHC Class I-Encoded Protein with the Potential to PresentN-Myristoylated 4-mer Peptides to T Cells.
J Immunol., 202, 2019
|
|
7CCS
 
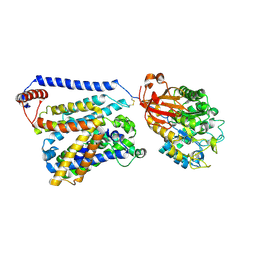 | | Consensus mutated xCT-CD98hc complex | | Descriptor: | 4F2 cell-surface antigen heavy chain, Consensus mutated Anionic Amino Acid Transporter Light Chain, Xc- System | | Authors: | Oda, K, Lee, Y, Takemoto, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-06-17 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Consensus mutagenesis approach improves the thermal stability of system x c - transporter, xCT, and enables cryo-EM analyses.
Protein Sci., 29, 2020
|
|
5B6S
 
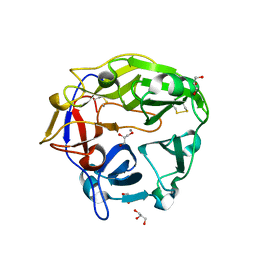 | | Catalytic domain of Coprinopsis cinerea GH62 alpha-L-arabinofuranosidase | | Descriptor: | CALCIUM ION, GLYCEROL, Glycosyl hydrolase family 62 protein | | Authors: | Tonozuka, T. | | Deposit date: | 2016-06-01 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Catalytic Domain of alpha-L-Arabinofuranosidase from Coprinopsis cinerea, CcAbf62A, Provides Insights into Structure-Function Relationships in Glycoside Hydrolase Family 62
Appl. Biochem. Biotechnol., 181, 2017
|
|
5B6T
 
 | | Catalytic domain of Coprinopsis cinerea GH62 alpha-L-arabinofuranosidase complexed with Pb | | Descriptor: | CALCIUM ION, GLYCEROL, Glycosyl hydrolase family 62 protein, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2016-06-01 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of the Catalytic Domain of alpha-L-Arabinofuranosidase from Coprinopsis cinerea, CcAbf62A, Provides Insights into Structure-Function Relationships in Glycoside Hydrolase Family 62
Appl. Biochem. Biotechnol., 181, 2017
|
|
6JJU
 
 | | Structure of Ca2+ ATPase | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Inoue, M, Sakuta, N, Watanabe, S, Inaba, K. | | Deposit date: | 2019-02-27 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Sarco/Endoplasmic Reticulum Ca2+-ATPase 2b Regulation via Transmembrane Helix Interplay.
Cell Rep, 27, 2019
|
|
7C24
 
 | | Glycosidase F290Y at pH8.0 | | Descriptor: | AMMONIUM ION, GLYCEROL, Isomaltose glucohydrolase | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C25
 
 | | Glycosidase Wild Type at pH8.0 | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C26
 
 | | Glycosidase Wild Type at pH4.5 | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C27
 
 | | Glycosidase F290Y at pH4.5 | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
2P6O
 
 | | Crystal structure of TTHB049 from Thermus thermophilus HB8 | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase | | Authors: | Yamamoto, H, Taketa, M, Tanaka, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-19 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of TTHB049 from Thermus thermophilus HB8
To be Published
|
|
2PCM
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | DIPHTHINE SYNTHASE, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yamamoto, H, Taketa, M, Tanaka, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
1WT5
 
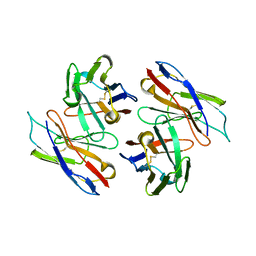 | | The Crystal Structure Of A Humanized Antibody Fv 528 | | Descriptor: | ANTI EGFR ANTIBODY FV REGION | | Authors: | Makabe, K, Tsumoto, K, Asano, R, Kondo, H, Kumagai, I. | | Deposit date: | 2004-11-16 | | Release date: | 2005-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermodynamic consequences of mutations in vernier zone residues of a humanized anti-human epidermal growth factor receptor murine antibody, 528
J.Biol.Chem., 283, 2008
|
|
8I90
 
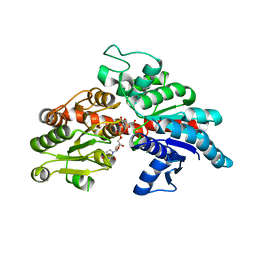 | |
8I94
 
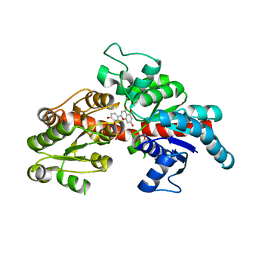 | | Structure of flavone 4'-O-glucoside 7-O-glucosyltransferase from Nemophila menziesii, complex with luteolin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, Glycosyltransferase, SULFATE ION | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Molecular basis of ligand recognition specificity of flavone glucosyltransferases in Nemophila menziesii.
Arch.Biochem.Biophys., 753, 2024
|
|
8I8Z
 
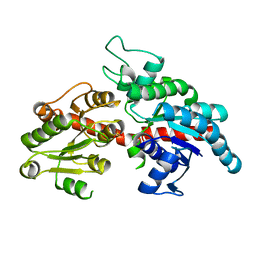 | |
5YO8
 
 | |
2EJJ
 
 | |
2E4R
 
 | |
2EJK
 
 | |
2EMU
 
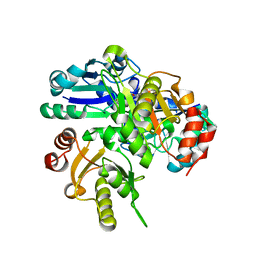 | |
2EN5
 
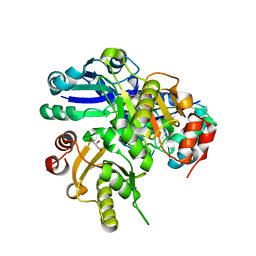 | |
2E4N
 
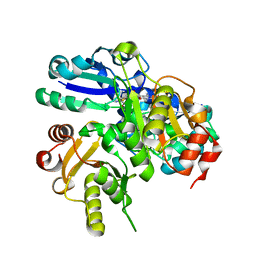 | |
2EH2
 
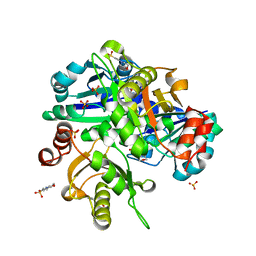 | |
6LA0
 
 | | Crystal structure of AoRut | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoside hydrolase family 5 | | Authors: | Koseki, T, Makabe, K. | | Deposit date: | 2019-11-11 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Aspergillus oryzae Rutinosidase: Biochemical and Structural Investigation.
Appl.Environ.Microbiol., 87, 2021
|
|
