2NLU
 
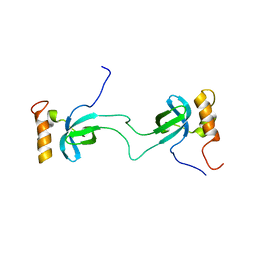 | |
2AJE
 
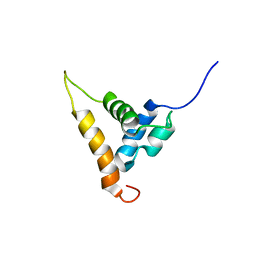 | | Solution structure of the Arabidopsis thaliana telomeric repeat-binding protein DNA binding domain | | Descriptor: | telomere repeat-binding protein | | Authors: | Hsiao, H.H, Sue, S.C, Chung, B.C, Cheng, Y.H, Huang, T.H. | | Deposit date: | 2005-08-01 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Arabidopsis thaliana telomeric repeat-binding protein DNA binding domain: a new fold with an additional C-terminal helix
J.Mol.Biol., 356, 2006
|
|
4QFQ
 
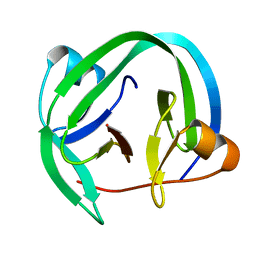 | |
8GUF
 
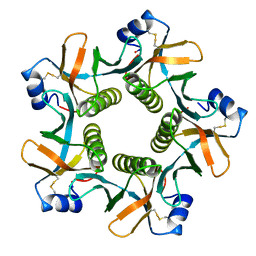 | |
8GW2
 
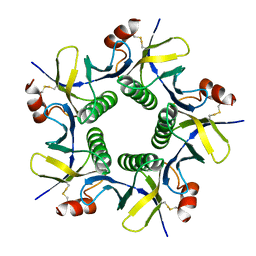 | |
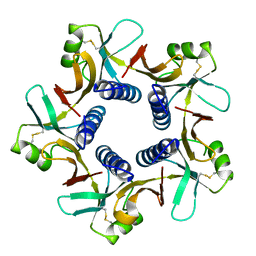
8H2R
 
 | |
6LOG
 
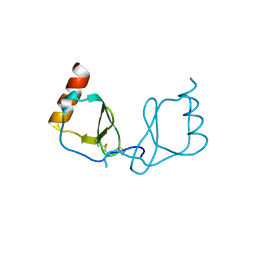 | |
4HSV
 
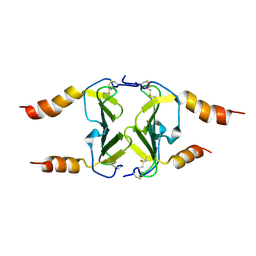 | | Crystal Structure of CXCL4L1 | | Descriptor: | Platelet factor 4 variant | | Authors: | Kuo, J.H, Liu, J.S, Wu, W.G, Sue, S.C. | | Deposit date: | 2012-10-30 | | Release date: | 2013-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.083 Å) | | Cite: | Alternative C-terminal helix orientation alters chemokine function: structure of the anti-angiogenic chemokine, CXCL4L1
J.Biol.Chem., 288, 2013
|
|
6AEZ
 
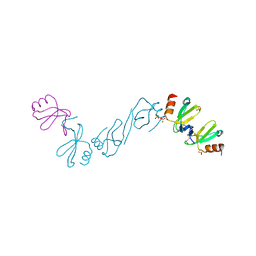 | | Crystal structure of human CCL5 trimer | | Descriptor: | C-C motif chemokine 5, SULFATE ION | | Authors: | Chen, Y.C, Li, K.M, Chen, P.J, Zarivach, R, Sun, Y.J, Sue, S.C. | | Deposit date: | 2018-08-07 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Integrative Model to Coordinate the Oligomerization and Aggregation Mechanisms of CCL5.
J.Mol.Biol., 432, 2020
|
|
1I02
 
 | |
5YAM
 
 | |
2CJR
 
 | |
