5ZWP
 
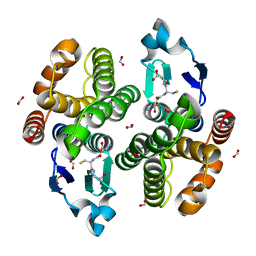 | |
3AIW
 
 | | Crystal structure of beta-glucosidase in rye complexed with 2-deoxy-2-fluoroglucoside and dinitrophenol | | Descriptor: | 2,4-DINITROPHENOL, 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucosidase | | Authors: | Sue, M, Nakamura, C, Miyamoto, T, Yajima, S. | | Deposit date: | 2010-05-18 | | Release date: | 2011-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active-site architecture of benzoxazinone-glucoside beta-D-glucosidases in Triticeae
Plant Sci., 180, 2011
|
|
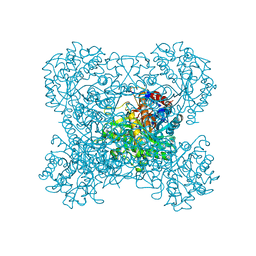
3AIV
 
 | | Crystal structure of beta-glucosidase in rye complexed with an aglycone DIMBOA | | Descriptor: | 2,4-DIHYDROXY-7-(METHYLOXY)-2H-1,4-BENZOXAZIN-3(4H)-ONE, Beta-glucosidase | | Authors: | Sue, M, Nakamura, C, Miyamoto, T, Yajima, S. | | Deposit date: | 2010-05-18 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active-site architecture of benzoxazinone-glucoside beta-D-glucosidases in Triticeae
Plant Sci., 180, 2011
|
|
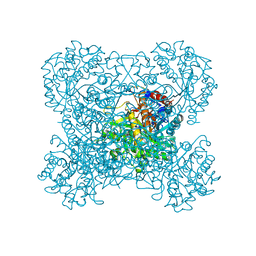
3AIU
 
 | | Crystal structure of beta-glucosidase in rye | | Descriptor: | Beta-glucosidase, GLYCEROL, SULFATE ION | | Authors: | Sue, M, Nakamura, C, Miyamoto, T, Yajima, S. | | Deposit date: | 2010-05-18 | | Release date: | 2011-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active-site architecture of benzoxazinone-glucoside beta-D-glucosidases in Triticeae
Plant Sci., 180, 2011
|
|
3AIR
 
 | | Crystal structure of beta-glucosidase in wheat complexed with 2-deoxy-2-fluoroglucoside and dinitrophenol | | Descriptor: | 2,4-DINITROPHENOL, 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucosidase | | Authors: | Sue, M, Nakamura, C, Miyamoto, T, Yajima, S. | | Deposit date: | 2010-05-18 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active-site architecture of benzoxazinone-glucoside beta-D-glucosidases in Triticeae
Plant Sci., 180, 2011
|
|
3AIS
 
 | | Crystal structure of a mutant beta-glucosidase in wheat complexed with DIMBOA-Glc | | Descriptor: | (2S)-2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one, Beta-glucosidase, beta-D-glucopyranose | | Authors: | Sue, M, Nakamura, C, Miyamoto, T, Yajima, S. | | Deposit date: | 2010-05-18 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active-site architecture of benzoxazinone-glucoside beta-D-glucosidases in Triticeae
Plant Sci., 180, 2011
|
|
3AIQ
 
 | | Crystal structure of beta-glucosidase in wheat complexed with an aglycone DIMBOA | | Descriptor: | 2,4-DIHYDROXY-7-(METHYLOXY)-2H-1,4-BENZOXAZIN-3(4H)-ONE, Beta-glucosidase | | Authors: | Sue, M, Nakamura, C, Miyamoto, T, Yajima, S. | | Deposit date: | 2010-05-18 | | Release date: | 2011-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active-site architecture of benzoxazinone-glucoside beta-D-glucosidases in Triticeae
Plant Sci., 180, 2011
|
|
2DGA
 
 | | Crystal structure of hexameric beta-glucosidase in wheat | | Descriptor: | Beta-glucosidase, GLYCEROL, SULFATE ION | | Authors: | Sue, M, Yamazaki, K, Miyamoto, T, Yajima, S. | | Deposit date: | 2006-03-10 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular and Structural Characterization of Hexameric beta-D-Glucosidases in Wheat and Rye.
Plant Physiol., 141, 2006
|
|
7DTP
 
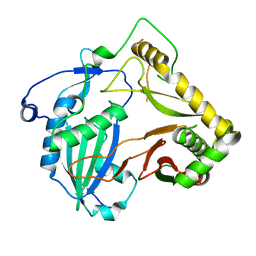 | | Crystal structure of agmatine coumaroyltransferase from Triticum aestivum | | Descriptor: | agmatine coumaroyltransferase | | Authors: | Yamane, M, Takenoya, M, Sue, M, Yajima, S. | | Deposit date: | 2021-01-06 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular and structural characterization of agmatine coumaroyltransferase in Triticeae, the key regulator of hydroxycinnamic acid amide accumulation.
Phytochemistry, 189, 2021
|
|
7CYS
 
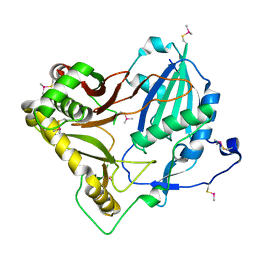 | | Crystal structure of barley agmatine coumaroyltransferase (HvACT), an N-acyltransferase in BAHD superfamily | | Descriptor: | Agmatine coumaroyltransferase-1 | | Authors: | Yamane, M, Takenoya, M, Sue, M, Yajima, S. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of barley agmatine coumaroyltransferase, an N-acyltransferase from the BAHD superfamily.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5YGV
 
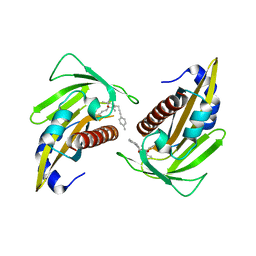 | | Crystal structure of the abscisic acid receptor PYR1 in complex with an antagonist | | Descriptor: | (2Z,4E)-3-methyl-5-[(1S,4S)-2,6,6-trimethyl-4-[3-(4-methylphenyl)prop-2-ynoxy]-1-oxidanyl-cyclohex-2-en-1-yl]penta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Akiyama, T, Sue, M, Takeuchi, J, Mimura, N, Okamoto, M, Monda, K, Iba, K, Ohnishi, T, Todoroki, Y, Yajima, S. | | Deposit date: | 2017-09-27 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Chemical Design of Abscisic Acid Antagonists That Block PYL-PP2C Receptor Interactions.
ACS Chem. Biol., 13, 2018
|
|
2H4O
 
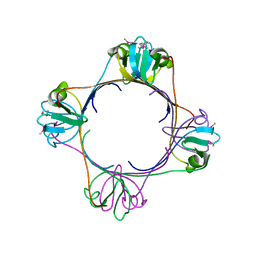 | | X-ray Crystal Structure of Protein yonK from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR415 | | Descriptor: | YonK protein | | Authors: | Seetharaman, J, Sue, M, Forouhar, F, Ken, C, Bonnie, C, Ma, L, Xiao, R, Acton, T.B, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-24 | | Release date: | 2006-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the hypothetical protein from bacillus subtilis (yonk).
To be Published
|
|
3WG8
 
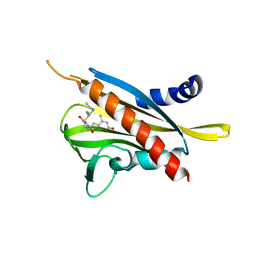 | | Crystal structure of the abscisic acid receptor PYR1 in complex with an antagonist AS6 | | Descriptor: | (2Z,4E)-5-[(1S)-3-(hexylsulfanyl)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Akiyama, T, Sue, M, Takeuchi, J, Okamoto, M, Muto, T, Endo, A, Nambara, E, Hirai, N, Ohnishi, T, Cutler, S.R, Todoroki, Y, Yajima, S. | | Deposit date: | 2013-07-31 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Designed abscisic acid analogs as antagonists of PYL-PP2C receptor interactions
Nat.Chem.Biol., 10, 2014
|
|
3VWX
 
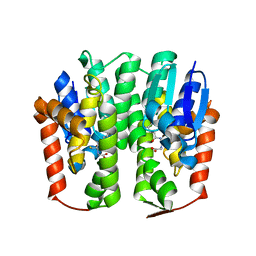 | | Structural analysis of an epsilon-class glutathione S-transferase from housefly, Musca domestica | | Descriptor: | GLUTATHIONE, Glutathione s-transferase 6B | | Authors: | Nakamura, C, Sue, M, Miyamoto, T, Yajima, S. | | Deposit date: | 2012-09-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of an epsilon-class glutathione transferase from housefly, Musca domestica
Biochem.Biophys.Res.Commun., 430, 2013
|
|
4X8Y
 
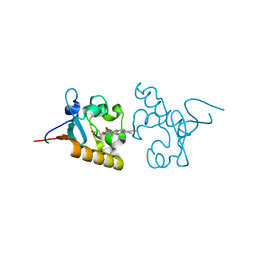 | | Crystal structure of human PGRMC1 cytochrome b5-like domain | | Descriptor: | Membrane-associated progesterone receptor component 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nakane, T, Yamamoto, T, Shimamura, T, Kobayashi, T, Kabe, Y, Suematsu, M. | | Deposit date: | 2014-12-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Haem-dependent dimerization of PGRMC1/Sigma-2 receptor facilitates cancer proliferation and chemoresistance
Nat Commun, 7, 2016
|
|
3VH9
 
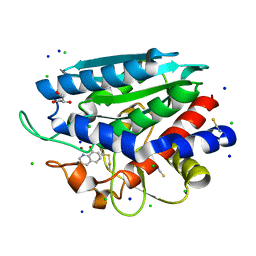 | | Crystal structure of Aeromonas proteolytica aminopeptidase complexed with 8-quinolinol | | Descriptor: | Bacterial leucyl aminopeptidase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saijo, S, Hanaya, K, Suetsugu, M, Kobayashi, K, Yamato, I, Aoki, S. | | Deposit date: | 2011-08-24 | | Release date: | 2012-05-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Potent inhibition of dinuclear zinc(II) peptidase, an aminopeptidase from Aeromonas proteolytica, by 8-quinolinol derivatives: inhibitor design based on Zn(2+) fluorophores, kinetic, and X-ray crystallographic study.
J.Biol.Inorg.Chem., 17, 2012
|
|
2YVA
 
 | | Crystal structure of Escherichia coli DiaA | | Descriptor: | DnaA initiator-associating protein diaA | | Authors: | Keyamura, K, Fujikawa, N, Ishida, T, Ozaki, S, Suetsugu, M, Kagawa, W, Yokoyama, S, Kurumizaka, H, Katayama, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-10 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes
Genes Dev., 21, 2007
|
|
6KB7
 
 | | X-ray structure of human PPARalpha ligand binding domain-Wy14643 co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KAY
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by soaking | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB3
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB2
 
 | | X-ray structure of human PPARalpha ligand binding domain-Wy14643 co-crystals obtained by soaking | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB9
 
 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by cross-seeding | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KAZ
 
 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by soaking | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB5
 
 | | X-ray structure of human PPARalpha ligand binding domain-5,8,11,14-eicosatetraynoic Acid (ETYA) co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, icosa-5,8,11,14-tetraynoic acid | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KBA
 
 | | X-ray structure of human PPARalpha ligand binding domain-Wy14643 co-crystals obtained by co-crystallization | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
