6LVC
 
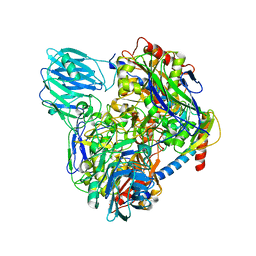 | | Structure of Dimethylformamidase, dimer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LVD
 
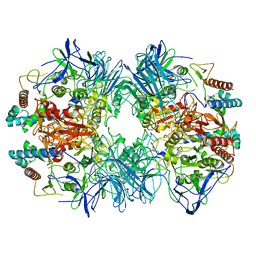 | | Structure of Dimethylformamidase, tetramer, Y440A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6MLR
 
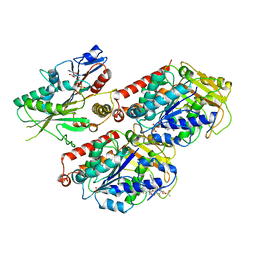 | | Cryo-EM structure of microtubule-bound Kif7 in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF7, ... | | Authors: | Mani, N, Jiang, S, Wilson-Kubalek, E.M, Ku, P, Milligan, R.A, Subramanian, R. | | Deposit date: | 2018-09-27 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Interplay between the Kinesin and Tubulin Mechanochemical Cycles Underlies Microtubule Tip Tracking by the Non-motile Ciliary Kinesin Kif7.
Dev.Cell, 49, 2019
|
|
6MLQ
 
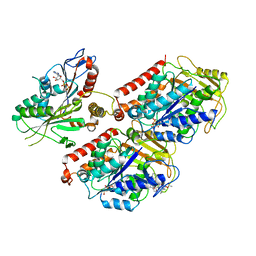 | | Cryo-EM structure of microtubule-bound Kif7 in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mani, N, Jiang, S, Wilson-Kubalek, E.M, Ku, P, Milligan, R.A, Subramanian, R. | | Deposit date: | 2018-09-27 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Interplay between the Kinesin and Tubulin Mechanochemical Cycles Underlies Microtubule Tip Tracking by the Non-motile Ciliary Kinesin Kif7.
Dev.Cell, 49, 2019
|
|
7W8J
 
 | | Dimethylformamidase, 2x(A2B2) | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Vinothkumar, K.R, Subramanian, R, Arya, C, Ramanathan, G. | | Deposit date: | 2021-12-07 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dimethylformamidase with a Unique Iron Center
To Be Published
|
|
5ZA4
 
 | |
5ZSX
 
 | | Catechol 2,3-dioxygenase with 3-fluorocatechol from Diaphorobacter sp DS2 | | Descriptor: | 1,2-ETHANEDIOL, 3-FLUOROBENZENE-1,2-DIOL, CALCIUM ION, ... | | Authors: | Mishra, K, Arya, C.K, Subramanian, R, Ramanathan, G. | | Deposit date: | 2018-04-30 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catechol 2,3-dioxygenase with 3-fluorocatechol from Diaphorobacter sp DS2.
To Be Published
|
|
5ZSZ
 
 | | Catechol 2,3-dioxygenase (C23O64) from Diaphorobacter sp DS2 | | Descriptor: | CALCIUM ION, Catechol 2,3-dioxygenase, Extradiol ring cleavage protein, ... | | Authors: | Mishra, K, Arya, C.K, Subramanian, R, Ramanathan, G. | | Deposit date: | 2018-04-30 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catechol 2,3-dioxygenase (C23O64) from Diaphorobacter sp DS2
To Be Published
|
|
8THI
 
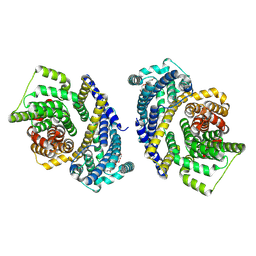 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (parallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, SODIUM ION, Sialic acid TRAP transporter permease protein SiaT | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
8THJ
 
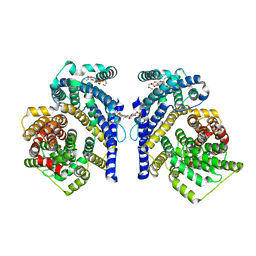 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (antiparallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
7D75
 
 | |
7VWW
 
 | |
6XT2
 
 | | EQADH-NADH-HEPTAFLUOROBUTANOL, P21 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2,2,3,3,4,4,4-heptafluorobutan-1-ol, ... | | Authors: | Plapp, B.V, Ramaswamy, S. | | Deposit date: | 2020-07-16 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Alternative binding modes in abortive NADH-alcohol complexes of horse liver alcohol dehydrogenase.
Arch.Biochem.Biophys., 701, 2021
|
|
6Q28
 
 | |
7Q02
 
 | | Zn-free structure of lipocalin-like Milk protein, inspired from Diploptera punctata, expressed in Saccharomyces cerevisiae | | Descriptor: | Milk protein, PALMITOLEIC ACID | | Authors: | Banerjee, S, Dhanabalan, K.V, Ramaswamy, S. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of recombinantly expressed cockroach Lili-Mip protein in glycosylated and deglycosylated forms.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
3S2A
 
 | | Crystal structure of PI3K-gamma in complex with a quinoline inhibitor | | Descriptor: | N-{2-chloro-5-[4-(morpholin-4-yl)quinolin-6-yl]pyridin-3-yl}-4-fluorobenzenesulfonamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phospshoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors: Discovery and Structure-Activity Relationships of a Series of Quinoline and Quinoxaline Derivatives.
J.Med.Chem., 54, 2011
|
|
6VVA
 
 | |
7MFS
 
 | |
7MFN
 
 | |
7MQT
 
 | |
6LVV
 
 | | N, N-dimethylformamidase | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, N,N-dimethylformamidase large subunit, ... | | Authors: | Arya, C.K, Ramaswamy, S, Kutti, R.V, Gurunath, R. | | Deposit date: | 2020-02-05 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7O3K
 
 | |
4F1S
 
 | | Crystal structure of human PI3K-gamma in complex with a pyridyl-triazine-sulfonamide inhibitor | | Descriptor: | N-(5-{[3-(4-amino-6-methyl-1,3,5-triazin-2-yl)-5-(tetrahydro-2H-pyran-4-yl)pyridin-2-yl]amino}-2-chloropyridin-3-yl)methanesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and structure-activity relationships of dual PI3K/mTOR inhibitors based on a 4-amino-6-methyl-1,3,5-triazine sulfonamide scaffold.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4FLH
 
 | | Crystal structure of human PI3K-gamma in complex with AMG511 | | Descriptor: | 4-(2-[(5-fluoro-6-methoxypyridin-3-yl)amino]-5-{(1R)-1-[4-(methylsulfonyl)piperazin-1-yl]ethyl}pyridin-3-yl)-6-methyl-1,3,5-triazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-06-14 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective Class I Phosphoinositide 3-Kinase Inhibitors: Optimization of a Series of Pyridyltriazines Leading to the Identification of a Clinical Candidate, AMG 511.
J.Med.Chem., 55, 2012
|
|
6Q26
 
 | |
