4CEW
 
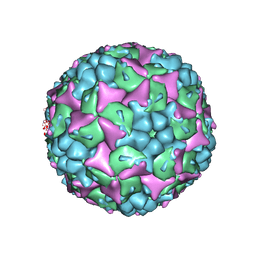 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor ALD | | Descriptor: | 4-[3-[(3s)-5-[4-[(e)-ethoxyiminomethyl]phenoxy]-3-methyl-pentyl]-2-oxidanylidene-imidazolidin-1-yl]pyridine-2-carboxamide, VP1, VP2, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDU
 
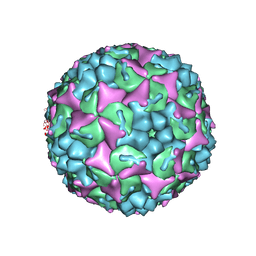 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CEY
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor NLD | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDX
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP12 | | Descriptor: | 1-(5-((3'-METHYL-[1,1'-BIPHENYL]-4-YL)OXY)PENTYL)-3-(, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-07 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDW
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP4 | | Descriptor: | 1-[(3S)-5-(4-iodanylphenoxy)-3-methyl-pentyl]-3-pyridin-4-yl-imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDQ
 
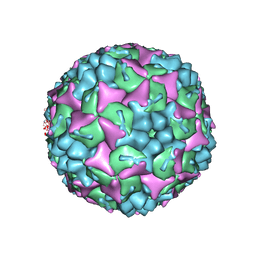 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP2 | | Descriptor: | 4-((5-(2-oxo-3-(pyridin-4-yl)imidazolidin-1-yl)pentyl)oxy)benzaldehyde O-ethyl oxime, SODIUM ION, VP1, ... | | Authors: | DeColibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
7TIC
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TI8
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TIB
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THV
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-12 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TKU
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TID
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THJ
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
1ZDJ
 
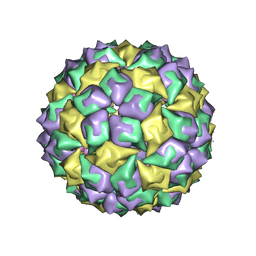 | | STRUCTURE OF BACTERIOPHAGE COAT PROTEIN-LOOP RNA COMPLEX | | Descriptor: | PROTEIN (MS2 PROTEIN CAPSID), RNA (5'-R(*GP*GP*AP*UP*CP*AP*CP*C)-3') | | Authors: | Grahn, E, Stonehouse, N, Valegard, K, Vandenworm, S, Liljas, L. | | Deposit date: | 1997-12-18 | | Release date: | 1998-07-08 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic studies of RNA hairpins in complexes with recombinant MS2 capsids: implications for binding requirements.
RNA, 5, 1999
|
|
1AQ3
 
 | | BACTERIOPHAGE MS2 CAPSID PROTEIN/RNA COMPLEX | | Descriptor: | PROTEIN (BACTERIOPHAGE MS2 COAT PROTEIN), RNA (5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*UP*AP*CP*CP*CP*AP*U P*GP*U)-3') | | Authors: | Van Den Worm, S, Stonehouse, N, Valegard, K, Liljas, L. | | Deposit date: | 1997-08-06 | | Release date: | 1997-12-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of MS2 coat protein mutants in complex with wild-type RNA operator fragments.
Nucleic Acids Res., 26, 1998
|
|
