1E4R
 
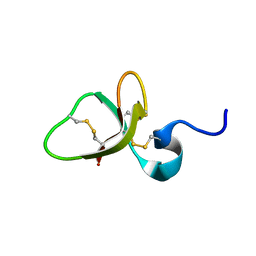 | | Solution structure of the mouse defensin mBD-8 | | Descriptor: | Beta-defensin 8 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity.
Protein Sci., 10, 2001
|
|
1E4S
 
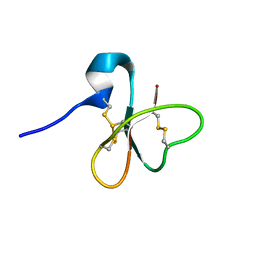 | | Solution structure of the human defensin hBD-1 | | Descriptor: | BETA-DEFENSIN 1 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of Human and Murine Beta-Defensins Reveals Structural Conservation in the Absence of Significant Sequence Similarity
Protein Sci., 10, 2001
|
|
1QFQ
 
 | |
1ROE
 
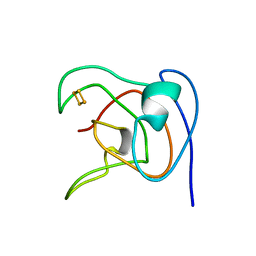 | | NMR STUDY OF 2FE-2S FERREDOXIN OF SYNECHOCOCCUS ELONGATUS | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Roesch, P, Baumann, B, Sticht, H, Sutter, M, Haehnel, W. | | Deposit date: | 1995-11-24 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Synechococcus elongatus [Fe2S2] ferredoxin in solution.
Biochemistry, 35, 1996
|
|
1TAC
 
 | |
1TBC
 
 | |
1WA7
 
 | | SH3 DOMAIN OF HUMAN LYN TYROSINE KINASE IN COMPLEX WITH A HERPESVIRAL LIGAND | | Descriptor: | HYPOTHETICAL 28.7 KDA PROTEIN IN DHFR 3'REGION (ORF1), TYROSINE-PROTEIN KINASE LYN | | Authors: | Bauer, F, Schweimer, K, Hoffmann, S, Roesch, P, Sticht, H. | | Deposit date: | 2004-10-25 | | Release date: | 2005-07-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigation of the Binding of a Herpesviral Protein to the SH3 Domain of Tyrosine Kinase Lck.
Biochemistry, 41, 2002
|
|
6T3X
 
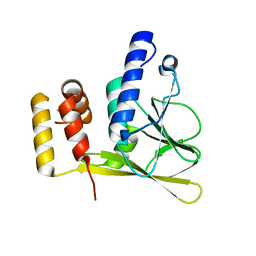 | | Crystal structure of the truncated human cytomegalovirus pUL50-pUL53 complex | | Descriptor: | Nuclear egress protein 2,Nuclear egress protein 1 | | Authors: | Muller, Y.A. | | Deposit date: | 2019-10-11 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | High-resolution crystal structures of two prototypical beta- and gamma-herpesviral nuclear egress complexes unravel the determinants of subfamily specificity.
J.Biol.Chem., 295, 2020
|
|
7PAB
 
 | | Varicella zoster Orf24-Orf27 nuclear egress complex | | Descriptor: | Nuclear egress protein 2,Nuclear egress protein 1, SULFATE ION, ZINC ION | | Authors: | Schweininger, J, Muller, Y.A. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the varicella-zoster Orf24-Orf27 nuclear egress complex spotlights multiple determinants of herpesvirus subfamily specificity.
J.Biol.Chem., 298, 2022
|
|
2KB6
 
 | | Solution structure of onconase C87A/C104A | | Descriptor: | Protein P-30 | | Authors: | Weininger, U, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Balbach, J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-11-24 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
5D5N
 
 | | Crystal Structure of the Human Cytomegalovirus pUL50-pUL53 Complex | | Descriptor: | Virion egress protein UL31 homolog, Virion egress protein UL34 homolog, ZINC ION | | Authors: | Walzer, S.A, Egerer-Sieber, C, Hohl, K, Sevvana, M, Muller, Y.A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structure of the Human Cytomegalovirus pUL50-pUL53 Core Nuclear Egress Complex Provides Insight into a Unique Assembly Scaffold for Virus-Host Protein Interactions.
J.Biol.Chem., 290, 2015
|
|
6T3Z
 
 | | Crystal structure of the truncated EBV BFRF1-BFLF2 nuclear egress complex | | Descriptor: | MALONATE ION, Nuclear egress protein 2,Nuclear egress protein 1 | | Authors: | Muller, Y.A. | | Deposit date: | 2019-10-11 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55862784 Å) | | Cite: | High-resolution crystal structures of two prototypical beta- and gamma-herpesviral nuclear egress complexes unravel the determinants of subfamily specificity.
J.Biol.Chem., 295, 2020
|
|
4WID
 
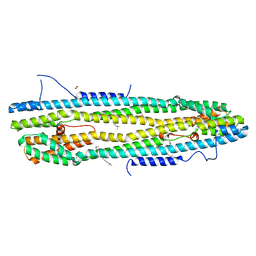 | | Crystal structure of the immediate-early 1 protein (IE1) at 2.31 angstrom (tetragonal form after crystal dehydration) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RhUL123 | | Authors: | Klingl, S, Scherer, M, Sevvana, M, Muller, Y.A, Stamminger, T. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Cytomegalovirus IE1 Protein Reveals Targeting of TRIM Family Member PML via Coiled-Coil Interactions.
Plos Pathog., 10, 2014
|
|
4OT1
 
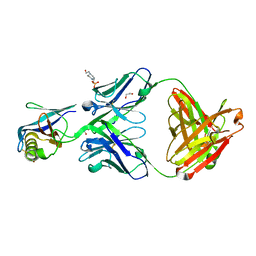 | |
4OSU
 
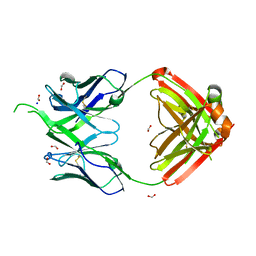 | |
3FD7
 
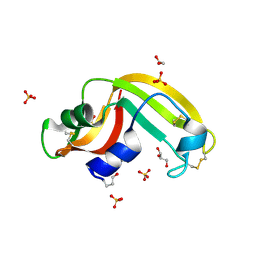 | | Crystal structure of Onconase C87A/C104A-ONC | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein P-30, ... | | Authors: | Neumann, P, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Stubbs, M.T. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
2NCN
 
 | |
2IIM
 
 | | SH3 Domain of Human Lck | | Descriptor: | CALCIUM ION, Proto-oncogene tyrosine-protein kinase LCK, TETRAETHYLENE GLYCOL, ... | | Authors: | Romir, J, Egerer-Sieber, C, Muller, Y.A. | | Deposit date: | 2006-09-28 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure analysis and solution studies of human Lck-SH3; zinc-induced homodimerization competes with the binding of proline-rich motifs.
J.Mol.Biol., 365, 2007
|
|
5MJ0
 
 | |
5MJ1
 
 | | Extracellular domain of human CD83 - rhombohedral crystal form | | Descriptor: | CD83 antigen, DI(HYDROXYETHYL)ETHER | | Authors: | Klingl, S, Egerer-Sieber, C, Schmid, B, Weiler, S, Muller, Y.A. | | Deposit date: | 2016-11-29 | | Release date: | 2017-03-29 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Extracellular Domain of the Human Dendritic Cell Surface Marker CD83.
J. Mol. Biol., 429, 2017
|
|
5MJ2
 
 | | Extracellular domain of human CD83 - rhombohedral crystal form after UV-RIP (S-SAD data) | | Descriptor: | CD83 antigen, DI(HYDROXYETHYL)ETHER | | Authors: | Klingl, S, Egerer-Sieber, C, Schmid, B, Weiler, S, Muller, Y.A. | | Deposit date: | 2016-11-29 | | Release date: | 2017-03-29 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of the Extracellular Domain of the Human Dendritic Cell Surface Marker CD83.
J. Mol. Biol., 429, 2017
|
|
5MIX
 
 | |
1HPH
 
 | |
1PFD
 
 | | THE SOLUTION STRUCTURE OF HIGH PLANT PARSLEY [2FE-2S] FERREDOXIN, NMR, 18 STRUCTURES | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Im, S.-C, Liu, G, Luchinat, C, Sykes, A.G, Bertini, I. | | Deposit date: | 1998-05-05 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of parsley [2Fe-2S]ferredoxin.
Eur.J.Biochem., 258, 1998
|
|
1TVT
 
 | |
