7CBC
 
 | |
5KOB
 
 | |
3KRS
 
 | |
4F4F
 
 | |
4F4A
 
 | |
4F0L
 
 | |
5VID
 
 | |
5VLI
 
 | | Computationally designed inhibitor peptide HB1.6928.2.3 in complex with influenza hemagglutinin (A/PuertoRico/8/1934) | | Descriptor: | 2,5,8,11-TETRAOXATRIDECANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Massively parallel de novo protein design for targeted therapeutics.
Nature, 550, 2017
|
|
3TMG
 
 | |
4H4G
 
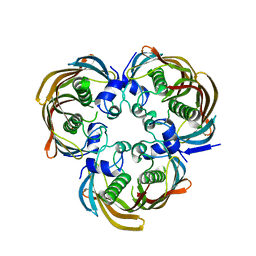 | | Crystal Structure of (3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase from Burkholderia thailandensis E264 | | Descriptor: | (3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase | | Authors: | Craig, T.K, Edwards, T.E, Staker, B, Stewart, L, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-09-17 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
6E9Z
 
 | | DHF119 filament | | Descriptor: | DHF119 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6E9T
 
 | | DHF58 filament | | Descriptor: | DHF58 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6E9Y
 
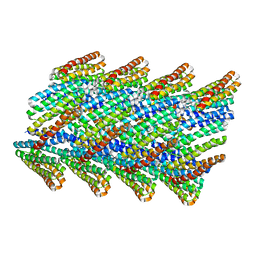 | | DHF38 filament | | Descriptor: | DHF38 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6E9R
 
 | | DHF46 filament | | Descriptor: | DHF46 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6E9X
 
 | | DHF91 filament | | Descriptor: | DHF91 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6E9V
 
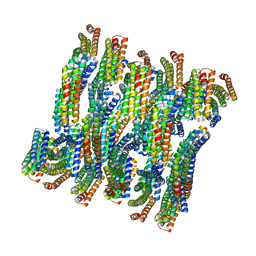 | | DHF79 filament | | Descriptor: | DHF79 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
8GJI
 
 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | GCG binder, Glucagon | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8GJG
 
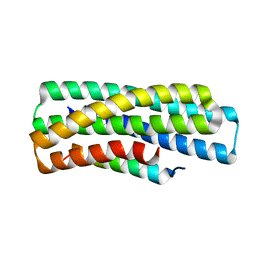 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | gluc_A04_0005, gluc_A04_0005 Binder | | Authors: | Leung, P.J.Y, Bera, A.K, Torres, S.V, Baker, D, Kang, A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8GLT
 
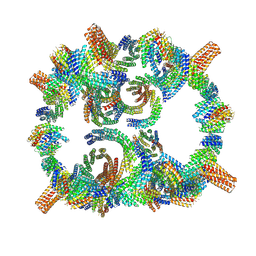 | | Backbone model of de novo-designed chlorophyll-binding nanocage O32-15 | | Descriptor: | C2-chlorophyll-comp_O32-15_ctermHis, polyalanine model, C3-comp_O32-15 | | Authors: | Redler, R.L, Ennist, N.M, Wang, S, Baker, D, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | De novo design of energy transfer proteins housing excitonically coupled chlorophyll special pairs
To Be Published
|
|
7N3T
 
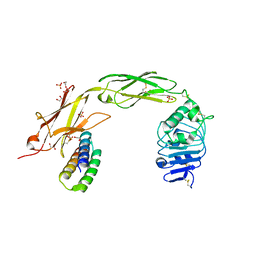 | | TrkA ECD complex with designed miniprotein ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jude, K.M, Cao, L, Garcia, K.C. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
8CTO
 
 | |
8CUN
 
 | |
8CWA
 
 | |
6DG5
 
 | | Structure of a de novo designed Interleukin-2/Interleukin-15 mimetic complex with IL-2Rb and IL-2Rg | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | Authors: | Jude, K.M, Silva, D.-A, Yu, S, Baker, D, Garcia, K.C. | | Deposit date: | 2018-05-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.516 Å) | | Cite: | De novo design of potent and selective mimics of IL-2 and IL-15.
Nature, 565, 2019
|
|
7N1J
 
 | |
