1CGP
 
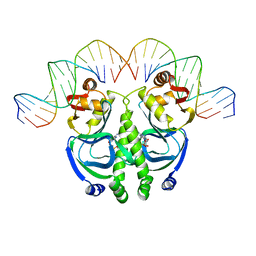 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*GP*TP*GP*TP*GP*AP*CP*AP*TP*AP*T)-3'), DNA (5'-D(*GP*TP*CP*AP*CP*AP*CP*TP*TP*TP*TP*CP*G)-3'), ... | | Authors: | Schultz, S.C, Shields, G.C, Steitz, T.A. | | Deposit date: | 1991-08-12 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees.
Science, 253, 1991
|
|
1DFU
 
 | |
1EFG
 
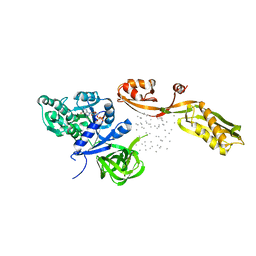 | | THE CRYSTAL STRUCTURE OF ELONGATION FACTOR G COMPLEXED WITH GDP, AT 2.7 ANGSTROMS RESOLUTION | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Czworkowski, J, Wang, J, Steitz, T.A, Moore, P.B. | | Deposit date: | 1994-10-17 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
EMBO J., 13, 1994
|
|
1FFY
 
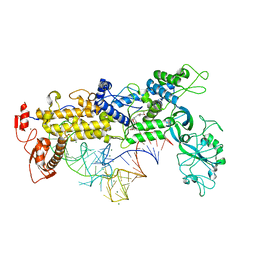 | | INSIGHTS INTO EDITING FROM AN ILE-TRNA SYNTHETASE STRUCTURE WITH TRNA(ILE) AND MUPIROCIN | | Descriptor: | ISOLEUCYL-TRNA, ISOLEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, ... | | Authors: | Silvian, L.F, Wang, J, Steitz, T.A. | | Deposit date: | 2000-07-26 | | Release date: | 2000-08-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin.
Science, 285, 1999
|
|
1GDT
 
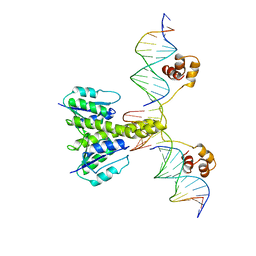 | |
1GPC
 
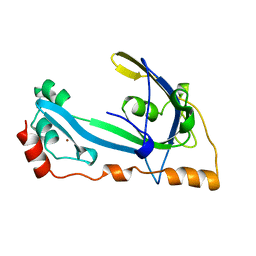 | | CORE GP32, DNA-BINDING PROTEIN | | Descriptor: | PROTEIN (CORE GP32), ZINC ION | | Authors: | Shamoo, Y, Friedman, A.M, Parsons, M.R, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1995-06-01 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA.
Nature, 376, 1995
|
|
1GTR
 
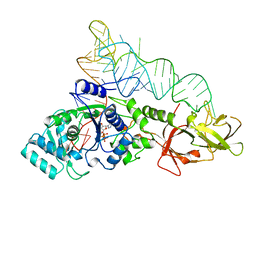 | |
1HA1
 
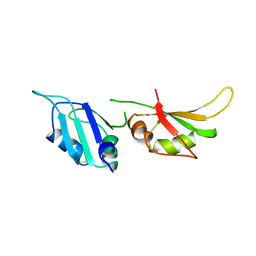 | | HNRNP A1 (RBD1,2) FROM HOMO SAPIENS | | Descriptor: | HNRNP A1 | | Authors: | Shamoo, Y, Krueger, U, Rice, L, Williams, K.R, Steitz, T.A. | | Deposit date: | 1996-10-30 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 A resolution.
Nat.Struct.Biol., 4, 1997
|
|
397D
 
 | |
3CB4
 
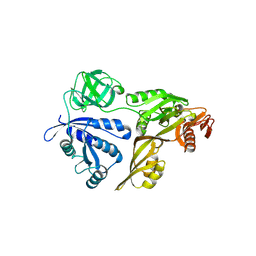 | | The Crystal Structure of LepA | | Descriptor: | GTP-binding protein lepA | | Authors: | Evans, R.N, Blaha, G, Bailey, S, Steitz, T.A. | | Deposit date: | 2008-02-21 | | Release date: | 2008-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of LepA, the ribosomal back translocase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CME
 
 | |
3CMA
 
 | |
3CPW
 
 | | The structure of the antibiotic LINEZOLID bound to the large ribosomal subunit of HALOARCULA MARISMORTUI | | Descriptor: | 23S RIBOSOMAL RNA, 5'-R(*CP*CP*AP*(PHE)*(ACA))-3', 50S ribosomal protein L10E, ... | | Authors: | Ippolito, J.A, Kanyo, Z.K, Wang, D, Franceschi, F.J, Moore, P.B, Steitz, T.A, Duffy, E.M. | | Deposit date: | 2008-04-01 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Oxazolidinone Antibiotic
Linezolid Bound to the 50S Ribosomal Subunit
J.Med.Chem., 51, 2008
|
|
3E0D
 
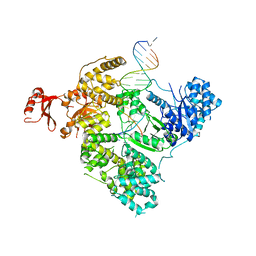 | | Insights into the Replisome from the Crystral Structure of the Ternary Complex of the Eubacterial DNA Polymerase III alpha-subunit | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase III subunit alpha, ... | | Authors: | Wing, R.A, Bailey, S, Steitz, T.A. | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Insights into the Replisome from the Structure of a Ternary Complex of the DNA Polymerase III alpha-Subunit.
J.Mol.Biol., 382, 2008
|
|
3E3J
 
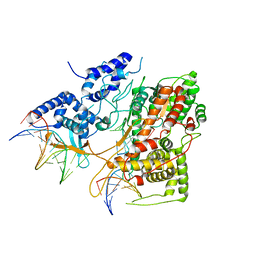 | | Crystal Structure of an Intermediate Complex of T7 RNAP and 8nt of RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*DTP*DAP*DAP*DTP*DAP*DCP*DGP*DAP*DCP*DTP*DCP*DAP*DCP*DTP*DAP*DTP*DAP*DTP*DTP*DTP*DCP*DTP*DGP*DCP*DCP*DAP*DAP*DAP*DCP*DGP*DGP*DC)-3'), DNA-directed RNA polymerase, ... | | Authors: | Durniak, K.J, Bailey, S, Steitz, T.A. | | Deposit date: | 2008-08-07 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (6.7 Å) | | Cite: | The structure of a transcribing t7 RNA polymerase in transition from initiation to elongation
Science, 322, 2008
|
|
3E2E
 
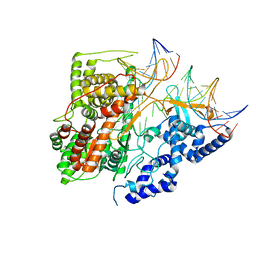 | | Crystal Structure of an Intermediate Complex of T7 RNAP and 7nt of RNA | | Descriptor: | DNA (28-MER), DNA (31-MER), DNA-directed RNA polymerase, ... | | Authors: | Durniak, K.J, Bailey, S, Steitz, T.A. | | Deposit date: | 2008-08-05 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of a transcribing t7 RNA polymerase in transition from initiation to elongation
Science, 322, 2008
|
|
3EDU
 
 | | Crystal structure of the ankyrin-binding domain of human erythroid spectrin | | Descriptor: | Spectrin beta chain, erythrocyte | | Authors: | Simonovic, M, Stabach, P, Simonovic, I, Steitz, T.A, Morrow, J.S. | | Deposit date: | 2008-09-03 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the ankyrin-binding site of {beta}-spectrin reveals how tandem spectrin-repeats generate unique ligand-binding properties
Blood, 113, 2009
|
|
1TAU
 
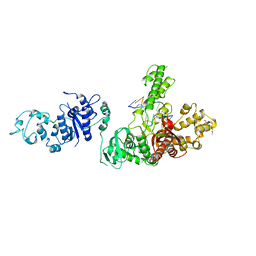 | | TAQ POLYMERASE (E.C.2.7.7.7)/DNA/B-OCTYLGLUCOSIDE COMPLEX | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, DNA (5'-D(*CP*GP*GP*AP*TP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*AP*TP*CP*CP*G)-3'), ... | | Authors: | Eom, S.H, Wang, J, Steitz, T.A. | | Deposit date: | 1996-06-17 | | Release date: | 1997-04-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Taq ploymerase with DNA at the polymerase active site.
Nature, 382, 1996
|
|
1TAQ
 
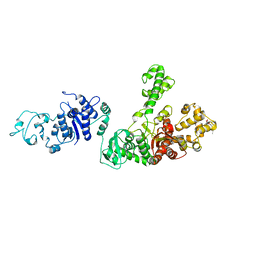 | | STRUCTURE OF TAQ DNA POLYMERASE | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, TAQ DNA POLYMERASE, ZINC ION | | Authors: | Kim, Y, Eom, S.H, Wang, J, Lee, D.-S, Suh, S.W, Steitz, T.A. | | Deposit date: | 1996-06-04 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Thermus aquaticus DNA polymerase.
Nature, 376, 1995
|
|
1VQN
 
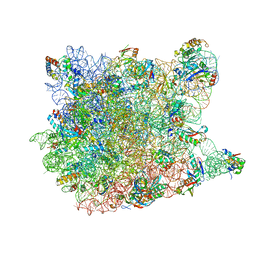 | |
1VQ6
 
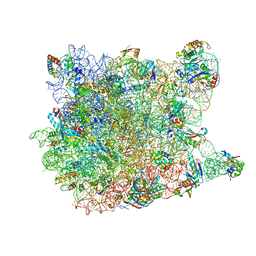 | |
1VQ7
 
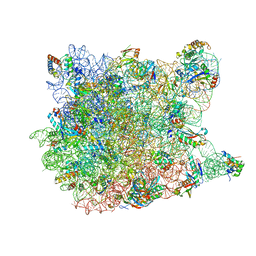 | |
2PZS
 
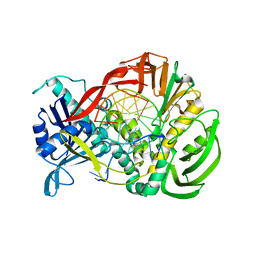 | | Phi29 DNA polymerase complexed with primer-template DNA (post-translocation binary complex) | | Descriptor: | 5'-d(CTAACACGTAAGCAGTC)-3', 5'-d(GACTGCTTAC)-3', DNA polymerase | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-18 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2PY5
 
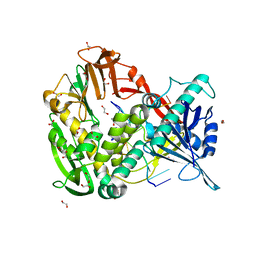 | | Phi29 DNA polymerase complexed with single-stranded DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-d(GGACTTT)-3', DNA polymerase | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-15 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2PFF
 
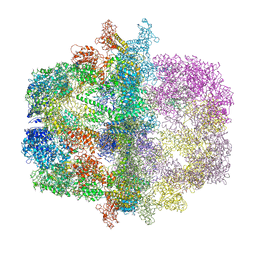 | | Structural Insights of Yeast Fatty Acid Synthase | | Descriptor: | Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, Tail protein | | Authors: | Xiong, Y, Lomakin, I.B, Steitz, T.A. | | Deposit date: | 2007-04-04 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Insights of Yeast Fatty Acid Synthase
Cell(Cambridge,Mass.), 129, 2007
|
|
