8FI5
 
 | |
8FI6
 
 | |
8FI4
 
 | |
8G0V
 
 | |
8G0R
 
 | |
8G0T
 
 | |
8G0U
 
 | |
8G0S
 
 | |
6MB0
 
 | |
6MB1
 
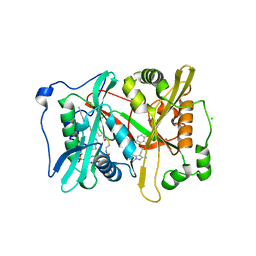 | | Crystal structure of N-myristoyl transferase (NMT) from Plasmodium vivax in complex with inhibitor IMP-1002 | | Descriptor: | 1,2-ETHANEDIOL, 1-(5-{4-fluoro-2-[2-(1,3,5-trimethyl-1H-pyrazol-4-yl)ethoxy]phenyl}-1-methyl-1H-indazol-3-yl)-N,N-dimethylmethanamine, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-08-29 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Identification of Resistance Breaking Antimalarial N‐Myristoyltransferase Inhibitors.
Cell Chem Biol, 26, 2019
|
|
6MAZ
 
 | |
6MAY
 
 | |
5IEQ
 
 | |
5IF2
 
 | |
5IFD
 
 | |
5IF5
 
 | |
5IFC
 
 | |
5IF8
 
 | |
5IFB
 
 | |
5IF7
 
 | |
5JMA
 
 | | Crystal structure of Mycobacterium avium SerB2 in complex with serine at catalytic (PSP) domain | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Phosphoserine phosphatase, ... | | Authors: | Shree, S, Dubey, S, Agrawal, A, Ramachandran, R. | | Deposit date: | 2016-04-28 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Mycobacterium avium SerB2 in complex with serine at catalytic (PSP) domain
To be published
|
|
5JLP
 
 | |
6AUB
 
 | |
6AUA
 
 | |
4IUJ
 
 | |
