2LGV
 
 | | Rbx1 | | Descriptor: | E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Spratt, D.E, Shaw, G.S. | | Deposit date: | 2011-08-02 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Selective recruitment of an e2~ubiquitin complex by an e3 ubiquitin ligase.
J.Biol.Chem., 287, 2012
|
|
2M9Y
 
 | |
2M48
 
 | | Solution Structure of IBR-RING2 Tandem Domain from Parkin | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE PARKIN, ZINC ION | | Authors: | Noh, Y.J, Mercier, P, Spratt, D.E, Shaw, G.S. | | Deposit date: | 2013-01-30 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A molecular explanation for the recessive nature of parkin-linked Parkinson's disease.
Nat Commun, 4, 2013
|
|
2LWR
 
 | | Solution Structure of RING2 Domain from Parkin | | Descriptor: | SD01679p, ZINC ION | | Authors: | Mercier, P, Spratt, D.E, Manczyk, N, Shaw, G.S. | | Deposit date: | 2012-08-06 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A molecular explanation for the recessive nature of parkin-linked Parkinson's disease.
Nat Commun, 4, 2013
|
|
5W87
 
 | |
8F36
 
 | |
8F37
 
 | |
2LL7
 
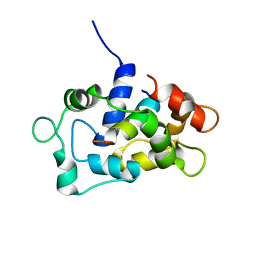 | | Solution NMR structure of CaM bound to the eNOS CaM binding domain peptide | | Descriptor: | Calmodulin, Nitric oxide synthase, endothelial | | Authors: | Piazza, M, Futrega, K, Spratt, D.E, Guillemette, J.G, Dieckmann, T. | | Deposit date: | 2011-10-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of Calmodulin (CaM) Bound to Nitric Oxide Synthase Peptides: Effects of a Phosphomimetic CaM Mutation.
Biochemistry, 51, 2012
|
|
5C23
 
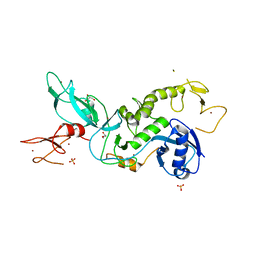 | | Parkin (S65DUblR0RBR) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Kumar, A, Aguirre, J.D, Condos, T.E.C, Martinez-Torres, R.J, Chaugule, V.K, Toth, R, Sundaramoorthy, R, Mercier, P, Knebel, A, Spratt, D.E, Barber, K.R, Shaw, G.S, Walden, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis.
Embo J., 34, 2015
|
|
5C1Z
 
 | | Parkin (UblR0RBR) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | kumar, A, Aguirre, J.D, Condos, T.E.C, Martinez-Torres, R.J, Chaugule, V.K, Toth, R, Sundaramoorthy, R, Mercier, P, Knebel, A, Spratt, D.E, Barber, K.R, Shaw, G.S, Walden, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis.
Embo J., 34, 2015
|
|
2LL6
 
 | | Solution NMR structure of CaM bound to iNOS CaM binding domain peptide | | Descriptor: | Calmodulin, Nitric oxide synthase, inducible | | Authors: | Piazza, M, Futrega, K, Spratt, D.E, Dieckmann, T, Guillemette, J.G. | | Deposit date: | 2011-10-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of Calmodulin (CaM) Bound to Nitric Oxide Synthase Peptides: Effects of a Phosphomimetic CaM Mutation.
Biochemistry, 51, 2012
|
|
6TGK
 
 | | Domain swapped E6AP C-lobe dimer | | Descriptor: | ACETATE ION, CALCIUM ION, Ubiquitin-protein ligase E3A | | Authors: | Ries, L.K, Feiler, C, Lowe, L.D, Liess, A.K.L, Lorenz, S. | | Deposit date: | 2019-11-16 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the catalytic C-lobe of the HECT-type ubiquitin ligase E6AP.
Protein Sci., 29, 2020
|
|
