1QE3
 
 | | PNB ESTERASE | | 分子名称: | PARA-NITROBENZYL ESTERASE, SULFATE ION, ZINC ION | | 著者 | Spiller, B, Gershenson, A, Arnold, F, Stevens, R. | | 登録日 | 1999-07-12 | | 公開日 | 1999-07-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A structural view of evolutionary divergence.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1C7I
 
 | | THERMOPHYLIC PNB ESTERASE | | 分子名称: | CALCIUM ION, PROTEIN (PARA-NITROBENZYL ESTERASE) | | 著者 | Spiller, B, Gershenson, A, Arnold, F, Stevens, R. | | 登録日 | 2000-02-21 | | 公開日 | 2000-03-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A structural view of evolutionary divergence.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1C7J
 
 | | PNB ESTERASE 56C8 | | 分子名称: | POTASSIUM ION, PROTEIN (PARA-NITROBENZYL ESTERASE), SULFATE ION | | 著者 | Spiller, B, Gershenson, A, Arnold, F, Stevens, R.C. | | 登録日 | 2000-02-21 | | 公開日 | 2000-03-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A structural view of evolutionary divergence.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3FCT
 
 | | MATURE METAL CHELATASE CATALYTIC ANTIBODY WITH HAPTEN | | 分子名称: | CADMIUM ION, CALCIUM ION, MAGNESIUM ION, ... | | 著者 | Romesberg, F.E, Santarsiero, B.D, Barnes, D, Yin, J, Spiller, B, Schultz, P.G, Stevens, R.C. | | 登録日 | 1999-06-13 | | 公開日 | 1999-06-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and kinetic evidence for strain in biological catalysis.
Biochemistry, 37, 1998
|
|
1A4J
 
 | | DIELS ALDER CATALYTIC ANTIBODY GERMLINE PRECURSOR | | 分子名称: | IMMUNOGLOBULIN, DIELS ALDER CATALYTIC ANTIBODY (HEAVY CHAIN), DIELS ALDER CATALYTIC ANTIBODY (LIGHT CHAIN) | | 著者 | Spiller, B.W, Romesburg, F.E, Schultz, P.G, Stevens, R.C. | | 登録日 | 1998-01-30 | | 公開日 | 1998-05-13 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Immunological origins of binding and catalysis in a Diels-Alderase antibody.
Science, 279, 1998
|
|
1A4K
 
 | | DIELS ALDER CATALYTIC ANTIBODY WITH TRANSITION STATE ANALOGUE | | 分子名称: | ANTIBODY FAB, CADMIUM ION, [4-(4-ACETYLAMINO-PHENYL)-3,5-DIOXO-4-AZA-TRICYCLO[5.2.2.0 2,6]UNDEC-1-YLCARBAMOYLOXY]-ACETIC ACID | | 著者 | Spiller, B.W, Romesburg, F.E, Schultz, P.G, Stevens, R.C. | | 登録日 | 1998-01-30 | | 公開日 | 1998-05-13 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Immunological origins of binding and catalysis in a Diels-Alderase antibody.
Science, 279, 1998
|
|
1UOT
 
 | | HUMAN CD55 DOMAINS 3 & 4 | | 分子名称: | COMPLEMENT DECAY-ACCELERATING FACTOR | | 著者 | Williams, P, Chaudhry, Y, Goodfellow, I.G, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | 登録日 | 2003-09-23 | | 公開日 | 2003-09-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
3HO6
 
 | |
3SRZ
 
 | | Clostridium difficile toxin A (TcdA) glucolsyltransferase domain bound to UDP-glucose | | 分子名称: | MANGANESE (II) ION, Toxin A, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | 著者 | Pruitt, R.N, Chumbler, N.M, Farrow, M.A, Seeback, S.A, Friedman, D.B, Spiller, B.W, Lacy, D.B. | | 登録日 | 2011-07-07 | | 公開日 | 2012-02-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structural Determinants of Clostridium difficile Toxin A Glucosyltransferase Activity.
J.Biol.Chem., 287, 2012
|
|
3SS1
 
 | | Clostridium difficile toxin A (TcdA) glucolsyltransferase domain | | 分子名称: | MANGANESE (II) ION, Toxin A | | 著者 | Pruitt, R.N, Chumbler, N.M, Farrow, M.A, Seeback, S.A, Friedman, D.B, Spiller, B.W, Lacy, D.B. | | 登録日 | 2011-07-07 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Structural Determinants of Clostridium difficile Toxin A Glucosyltransferase Activity.
J.Biol.Chem., 287, 2012
|
|
2QV3
 
 | |
1H04
 
 | | Human CD55 domains 3 & 4 | | 分子名称: | COMPLEMENT DECAY-ACCELERATING FACTOR, NICKEL (II) ION | | 著者 | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | 登録日 | 2002-06-11 | | 公開日 | 2003-03-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1H2P
 
 | | Human CD55 domains 3 & 4 | | 分子名称: | COMPLEMENT DECAY-ACCELERATING FACTOR | | 著者 | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | 登録日 | 2002-08-13 | | 公開日 | 2003-03-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1H03
 
 | | Human CD55 domains 3 & 4 | | 分子名称: | COMPLEMENT DECAY-ACCELERATING FACTOR | | 著者 | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | 登録日 | 2002-06-11 | | 公開日 | 2003-03-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
8G72
 
 | | SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A) | | 分子名称: | Nanosota-2, Spike glycoprotein | | 著者 | Ye, G, Bu, F, Liu, B, Li, F. | | 登録日 | 2023-02-16 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (5.6 Å) | | 主引用文献 | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
8G74
 
 | | SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | 著者 | Ye, G, Bu, F, Liu, B, Li, F. | | 登録日 | 2023-02-16 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
8G75
 
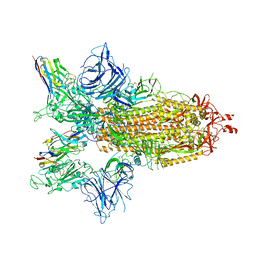 | | SARS-CoV-2 spike/Nb4 complex with 2 RBDs up and 3 Nb4 bound | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-4, ... | | 著者 | Ye, G, Bu, F, Liu, B, Li, F. | | 登録日 | 2023-02-16 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
8G73
 
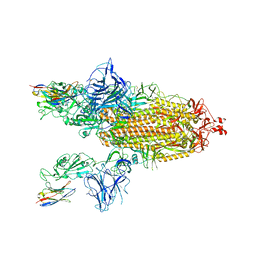 | | SARS-CoV-2 spike/Nb3 complex with 2 RBDs up and 3 Nb3 bound at 2.5 A | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | 著者 | Ye, G, Bu, F, Liu, B, Li, F. | | 登録日 | 2023-02-16 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
8G71
 
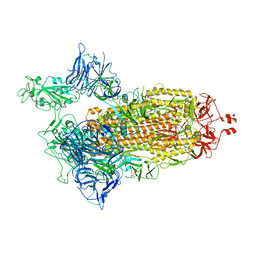 | | Spike/Nb2 complex with 1 RBD up | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Ye, G, Bu, F, Liu, B, Li, F. | | 登録日 | 2023-02-16 | | 公開日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | SARS-CoV-2 spike/Nb2 complex with 1 RBD up
To Be Published
|
|
8G79
 
 | |
8G7B
 
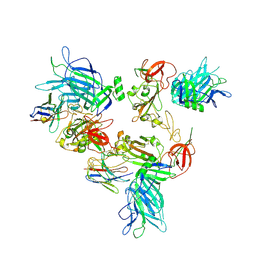 | |
8G76
 
 | | SARS-CoV-2 spike/Nb5 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-5, ... | | 著者 | Ye, G, Bu, F, Liu, B, Li, F. | | 登録日 | 2023-02-16 | | 公開日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | SARS-CoV-2 spike/Nb5 complex
To Be Published
|
|
8G77
 
 | | SARS-CoV-2 spike/Nb6 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-6, ... | | 著者 | Ye, G, Bu, F, Liu, B, Li, F. | | 登録日 | 2023-02-16 | | 公開日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | SARS-CoV-2 spike/Nb6 complex
To Be Published
|
|
8G7A
 
 | |
8G7C
 
 | |
