5QHF
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with NUOOA000301a | | Descriptor: | 2-(4-methoxyphenyl)-N-{5-[2-oxo-2-(3-oxopiperazin-1-yl)ethoxy]pyridin-3-yl}acetamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QHB
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with RK4-332 | | Descriptor: | 1-(4'-methyl[1,1'-biphenyl]-2-yl)pyrrolidin-2-one, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QGH
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with FMOPL000420a | | Descriptor: | (2R)-N-(1,2-oxazol-3-yl)-2-phenylbutanamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QGR
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with FMOPL000022a | | Descriptor: | 5-(pyridin-2-yl)thiophene-2-carbothioamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QH5
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with NUOOA000224a | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, N-(1,2-oxazol-3-yl)-2-phenylacetamide, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5N5G
 
 | |
5ACS
 
 | | Y233A-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | GIM-1 PROTEIN, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
5ACT
 
 | | W228S-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | GIM-1 PROTEIN, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
5ACQ
 
 | | W228A-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | BETA-LACTAMASE, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
5ACP
 
 | | W228R-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | GIM-1 PROTEIN, MAGNESIUM ION, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
5ACR
 
 | | W228Y-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | CALCIUM ION, GIM-1 PROTEIN, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
5EVK
 
 | | Crystal structure of the metallo-beta-lactamase L1 in complex with the bisthiazolidine inhibitor L-CS319 | | Descriptor: | (3R,5R,7aS)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.627 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EWA
 
 | | Crystal structure of the metallo-beta-lactamase IMP-1 in complex with the bisthiazolidine inhibitor L-VC26 | | Descriptor: | (3~{R},5~{R},7~{a}~{S})-2,2-dimethyl-5-(sulfanylmethyl)-3,5,7,7~{a}-tetrahydro-[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase IMP-1, ... | | Authors: | Kosmopoulou, M, Hinchliffe, P, Spencer, J. | | Deposit date: | 2015-11-20 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EVB
 
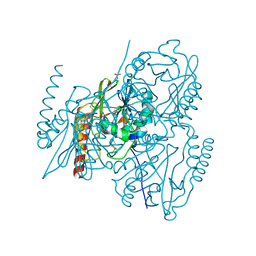 | | Crystal structure of the metallo-beta-lactamase L1 in complex with the bisthiazolidine inhibitor D-CS319 | | Descriptor: | (3S,5S,7aR)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EW0
 
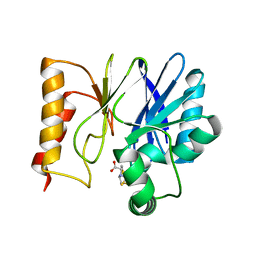 | | Crystal structure of the metallo-beta-lactamase Sfh-I in complex with the bisthiazolidine inhibitor L-CS319 | | Descriptor: | (3R,5R,7aS)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Beta-lactamase, ZINC ION | | Authors: | Hinchliffe, P, Tooke, C.L, Spencer, J. | | Deposit date: | 2015-11-20 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EVD
 
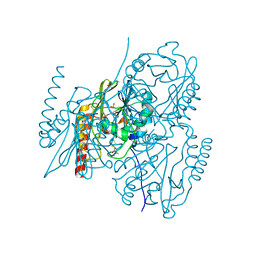 | | Crystal structure of the metallo-beta-lactamase L1 in complex with the bisthiazolidine inhibitor D-VC26 | | Descriptor: | (3S,5S,7aR)-2,2-dimethyl-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EV8
 
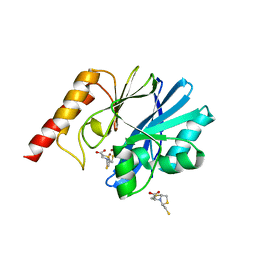 | | Crystal structure of the metallo-beta-lactamase IMP-1 in complex with the bisthiazolidine inhibitor D-CS319 | | Descriptor: | (3S,5S,7aR)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase IMP-1, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5NDB
 
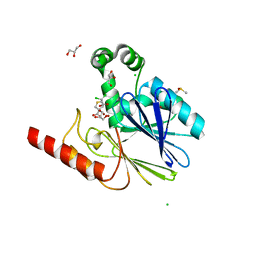 | | Crystal structure of metallo-beta-lactamase SPM-1 complexed with cyclobutanone inhibitor | | Descriptor: | (1~{S},4~{R},5~{S})-7,7-bis(chloranyl)-6,6-bis(oxidanyl)-2$l^{4}-thiabicyclo[3.2.0]hept-2-ene-4-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase IMP-1, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2017-03-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Cyclobutanone Mimics of Intermediates in Metallo-beta-Lactamase Catalysis.
Chemistry, 24, 2018
|
|
5NE3
 
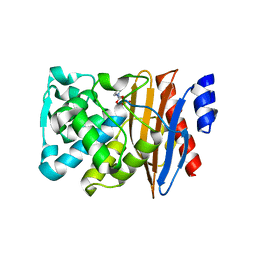 | | L2 class A serine-beta-lactamase complexed with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Hinchliffe, P, Calvopina, K, Spencer, J. | | Deposit date: | 2017-03-09 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural/mechanistic insights into the efficacy of nonclassical beta-lactamase inhibitors against extensively drug resistant Stenotrophomonas maltophilia clinical isolates.
Mol. Microbiol., 106, 2017
|
|
5NDE
 
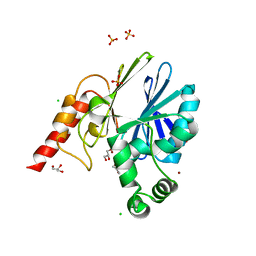 | |
5NE1
 
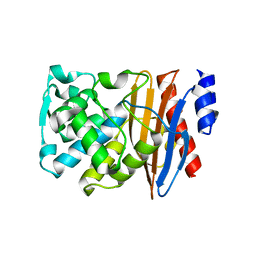 | | L2 class A serine-beta-lactamase in complex with cyclic boronate 2 | | Descriptor: | (4~{R})-4-[[4-(aminomethyl)phenyl]carbonylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, Beta-lactamase, TRIETHYLENE GLYCOL | | Authors: | Hinchliffe, P, Calvopina, K, Spencer, J. | | Deposit date: | 2017-03-09 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural/mechanistic insights into the efficacy of nonclassical beta-lactamase inhibitors against extensively drug resistant Stenotrophomonas maltophilia clinical isolates.
Mol. Microbiol., 106, 2017
|
|
5S8R
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | N-butyl-4-(furan-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassel-Hart , S, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal Structures of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 soaked with crude reaction mixtures
To Be Published
|
|
5S9J
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with starting material | | Descriptor: | 2-azanyl-3-methyl-benzoic acid, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassel-Hart , S, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structures of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 soaked with crude reaction mixtures
To Be Published
|
|
5S8S
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | N-(2-fluoro-4-methoxyphenyl)-4-(furan-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassel-Hart , S, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal Structures of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 soaked with crude reaction mixtures
To Be Published
|
|
5S8Z
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | N-[(3,4-dihydro-2H-1lambda~4~-thiophen-5-yl)methyl]-4-(furan-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassel-Hart , S, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structures of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 soaked with crude reaction mixtures
To Be Published
|
|
