2LOY
 
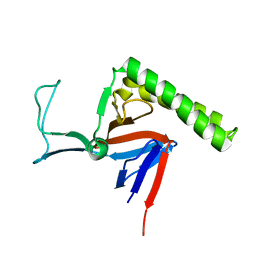 | | Refined Miminal Constraint Solution NMR Structure of Translationally-controlled tumor protein (TCTP) from Caenorhabditis elegans, Northeast Structural Genomics Consortium Target WR73 | | Descriptor: | Translationally-controlled tumor protein homolog | | Authors: | Aramini, J.M, Rossi, P, Cort, J.R, Lee, H, Janjua, H, Maglaqui, M, Cooper, B, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2ND3
 
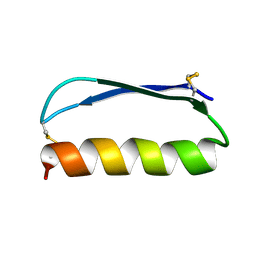 | | Solution structure of the de novo mini protein gEEH_04 | | Descriptor: | De novo mini protein EEH_04 | | Authors: | Pulavarti, S.V, Bahl, C.D, Gilmore, J.M, Eletsky, A, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2ND2
 
 | | Solution structure of the de novo mini protein gHHH_06 | | Descriptor: | De novo mini protein HHH_06 | | Authors: | Pulavarti, S.V, Eletsky, A, Bahl, C.D, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2LNU
 
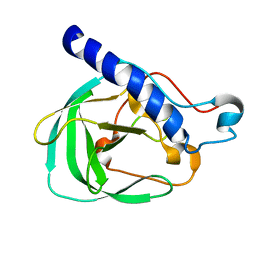 | | Solution NMR Structure of the uncharacterized protein from gene locus rrnAC0354 of Haloarcula marismortui, Northeast Structural Genomics Consortium Target HmR11 | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Liu, G, Lange, O.F, Lee, H, Janjua, H, Ciccosanti, C, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LOK
 
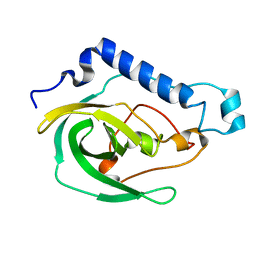 | | Solution NMR Structure of the uncharacterized protein from gene locus VNG_0733H of Halobacterium salinarium, Northeast Structural Genomics Consortium Target HsR50 | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Lange, O.F, Lee, H, Hamilton, K, Ciccosanti, C, Buchwald, W.A, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-24 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2KZN
 
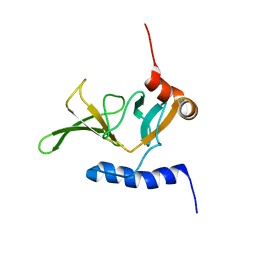 | | Solution NMR Structure of Peptide methionine sulfoxide reductase msrB from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR10 | | Descriptor: | Peptide methionine sulfoxide reductase msrB | | Authors: | Ertekin, A, Maglaqui, M, Janjua, H, Cooper, B, Ciccosanti, C, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J, Lee, H, Aramini, J.M, Rossi, P, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2Q1L
 
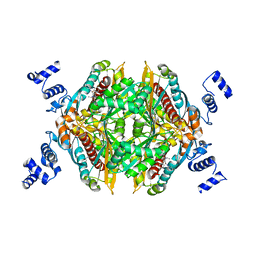 | | Design and Synthesis of Pyrrole-based, Hepatoselective HMG-CoA Reductase Inhibitors | | Descriptor: | (3R,5R)-7-[5-(ANILINOCARBONYL)-3,4-BIS(4-FLUOROPHENYL)-1-ISOPROPYL-1H-PYRROL-2-YL]-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Pfefferkorn, J.A, Harris, M.S, Finzel, B.C. | | Deposit date: | 2007-05-24 | | Release date: | 2007-07-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design and synthesis of hepatoselective, pyrrole-based HMG-CoA reductase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2Q6C
 
 | | Design and synthesis of novel, conformationally restricted HMG-COA reductase inhibitors | | Descriptor: | (3R,5R)-7-[1-(4-FLUOROPHENYL)-3-ISOPROPYL-4-OXO-5-PHENYL-4,5-DIHYDRO-3H-PYRROLO[2,3-C]QUINOLIN-2-YL]-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Pavlovsky, A, Pfefferkorn, J.A, Harris, M.S, Finzel, B.C. | | Deposit date: | 2007-06-04 | | Release date: | 2007-07-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of novel, conformationally restricted HMG-CoA reductase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2KW5
 
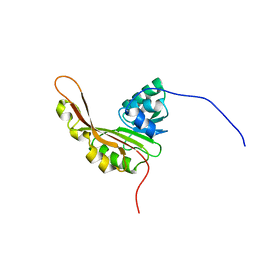 | | Solution NMR Structure of the Slr1183 protein from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR145 | | Descriptor: | Slr1183 protein | | Authors: | Rossi, P, Forouhar, F, Lee, H, Lange, O, Mao, B, Lemak, A, Maglaqui, M, Belote, R, Ciccosanti, C, Foote, E, Sahdev, S, Acton, T, Xiao, R, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2Q6B
 
 | | Design and synthesis of novel, conformationally restricted HMG-COA reductase inhibitors | | Descriptor: | (3R,5R)-7-[3-(4-FLUOROPHENYL)-1-ISOPROPYL-8-OXO-7-PHENYL-1,4,5,6,7,8-HEXAHYDROPYRROLO[2,3-C]AZEPIN-2-YL]-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Pavlovsky, A, Pfefferkorn, J.A, Harris, M.S, Finzel, B.C. | | Deposit date: | 2007-06-04 | | Release date: | 2007-07-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of novel, conformationally restricted HMG-CoA reductase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6LA5
 
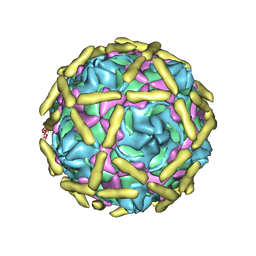 | |
6LB1
 
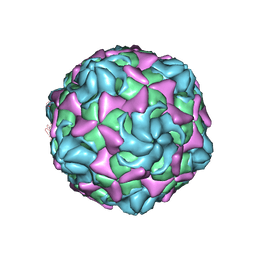 | | Cryo-EM structure of echovirus 11 A-particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-13 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
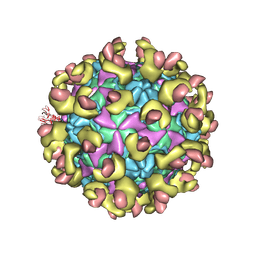
6LA7
 
 | |
6LA3
 
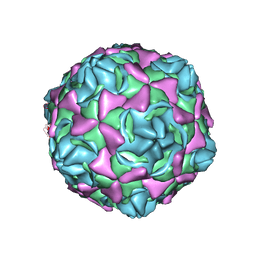 | | Cryo-EM structure of full echovirus 11 particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-11 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
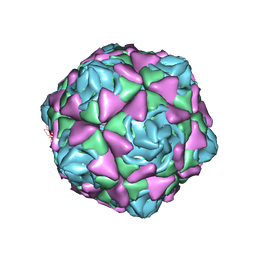
6LAO
 
 | |
6LBO
 
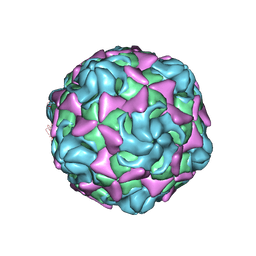 | | Cryo-EM structure of echovirus 11 empty particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LBQ
 
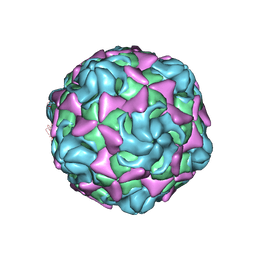 | | Cryo-EM structure of echovirus 11 empty particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LA4
 
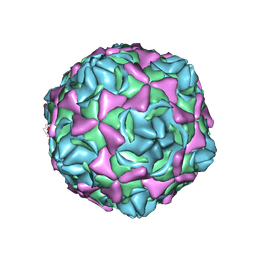 | | Cryo-EM structure of full echovirus 11 particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-11 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LA6
 
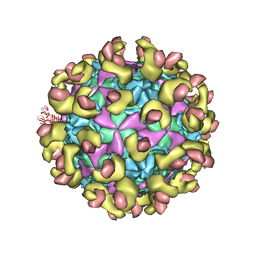 | |
6LAP
 
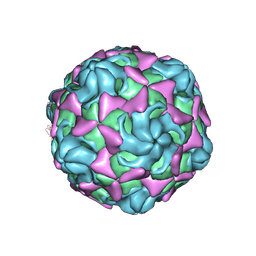 | | Cryo-EM structure of echovirus 11 A-particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-13 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LKX
 
 | | The structure of PRRSV helicase | | Descriptor: | CITRIC ACID, GLYCEROL, RNA-dependent RNA polymerase, ... | | Authors: | Shi, Y.J, Tong, X.H, Peng, G.Q. | | Deposit date: | 2019-12-20 | | Release date: | 2020-05-27 | | Last modified: | 2021-06-09 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Structural Characterization of the Helicase nsp10 Encoded by Porcine Reproductive and Respiratory Syndrome Virus.
J.Virol., 94, 2020
|
|
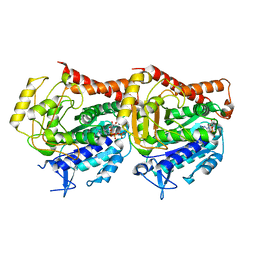
7YSP
 
 | |
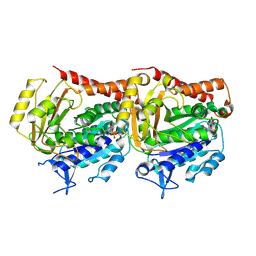
7YSO
 
 | |
7YSR
 
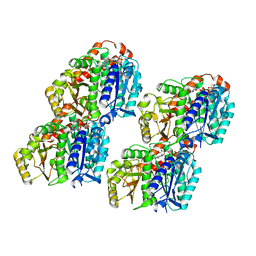 | | GTPgammaS MT decorated with kinesin | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Tubulin alpha chain, ... | | Authors: | Zhou, J, Wang, H.-W. | | Deposit date: | 2022-08-12 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into the mechanism of GTP initiation of microtubule assembly.
Nat Commun, 14, 2023
|
|
7YSN
 
 | | Tubulin heterodimer structure of GMPCPP state in solution | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Tubulin alpha-1B chain, ... | | Authors: | Zhou, J, Wang, H.-W. | | Deposit date: | 2022-08-12 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the mechanism of GTP initiation of microtubule assembly.
Nat Commun, 14, 2023
|
|
