1PLO
 
 | |
1KTZ
 
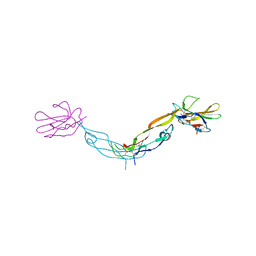 | | Crystal Structure of the Human TGF-beta Type II Receptor Extracellular Domain in Complex with TGF-beta3 | | Descriptor: | TGF-beta Type II Receptor, TRANSFORMING GROWTH FACTOR BETA 3 | | Authors: | Hart, P.J, Deep, S, Taylor, A.B, Shu, Z, Hinck, C.S, Hinck, A.P. | | Deposit date: | 2002-01-18 | | Release date: | 2002-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the human TbetaR2 ectodomain--TGF-beta3 complex.
Nat.Struct.Biol., 9, 2002
|
|
6ZDB
 
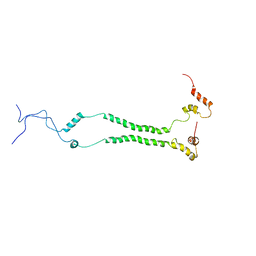 | | NMR structural analysis of yeast Cox13 reveals its C-terminus in interaction with ATP | | Descriptor: | Cytochrome c oxidase subunit 13, mitochondrial | | Authors: | Shu, Z, Pontus, P, Peter, B, Lena, M, Pia, A. | | Deposit date: | 2020-06-14 | | Release date: | 2021-05-19 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of the yeast cytochrome c oxidase subunit Cox13 and its interaction with ATP.
Bmc Biol., 19, 2021
|
|
7WN6
 
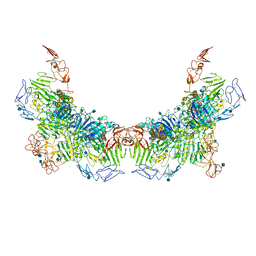 | | Cryo-EM structure of VWF D'D3 dimer (R1136M/E1143M mutant) complexed with D1D2 at 3.29 angstron resolution (2 units) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural mechanism of VWF D'D3 dimer formation.
Cell Discov, 8, 2022
|
|
7WN4
 
 | | Cryo-EM structure of VWF D'D3 dimer (wild type) complexed with D1D2 at 3.4 angstron resolution (1 unit) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of VWF D'D3 dimer formation.
Cell Discov, 8, 2022
|
|
7WN3
 
 | | Cryo-EM structure of VWF D'D3 dimer (2M mutant) complexed with D1D2 at 3.29 angstron resolution (2 units) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural mechanism of VWF D'D3 dimer formation.
Cell Discov, 8, 2022
|
|
7WPP
 
 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 2.85 angstron resolution (1 unit) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
7WPQ
 
 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 3.27 angstron resolution (2 units) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.267 Å) | | Cite: | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
7WQT
 
 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 4.3 angstron resolution (VWF tube) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-26 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
7WPS
 
 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 4.3 angstron resolution (7 units) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
7WPR
 
 | | VWF D'D3 dimer complexed with D1D2 at 4.39 angstron resolution(VWF tube) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
6KLS
 
 | | Hyperthermophilic respiratory Complex III | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, Cytochrome c, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guohong, P, Guoliang, Z, Hui, Z, Shuangbo, Z, Xiaoyun, P, Yan, Z. | | Deposit date: | 2019-07-30 | | Release date: | 2020-05-13 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6KLV
 
 | | Hyperthermophilic respiratory Complex III | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, ANTIMYCIN, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guohong, P, Guoliang, Z, Hui, Z, Shuangbo, Z, Xiaoyun, P, Yan, Z. | | Deposit date: | 2019-07-30 | | Release date: | 2020-05-13 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5Z1E
 
 | | MAP2K7 C218S mutant-inhibitor | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7, N-[3-(6-methyl-1H-indazol-3-yl)phenyl]prop-2-enamide | | Authors: | Kinoshita, T, London, N. | | Deposit date: | 2017-12-26 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Covalent Docking Identifies a Potent and Selective MKK7 Inhibitor.
Cell Chem Biol, 26, 2019
|
|
5Z1D
 
 | | MAP2K7 C276S mutant-inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Dual specificity mitogen-activated protein kinase kinase 7, N-[3-(6-methyl-1H-indazol-3-yl)phenyl]prop-2-enamide | | Authors: | Kinoshita, T, London, N. | | Deposit date: | 2017-12-26 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Covalent Docking Identifies a Potent and Selective MKK7 Inhibitor.
Cell Chem Biol, 26, 2019
|
|
5X56
 
 | | Crystal structure of PSB27 from Arabidopsis thaliana | | Descriptor: | Photosystem II repair protein PSB27-H1, chloroplastic | | Authors: | Liu, J, Cheng, X. | | Deposit date: | 2017-02-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Psb27 from Arabidopsis thaliana determined at a resolution of 1.85 angstrom.
Photosyn. Res., 136, 2018
|
|
6VAJ
 
 | | Crystal Structure Analysis of human PIN1 | | Descriptor: | 2-chloro-N-(2,2-dimethylpropyl)-N-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]acetamide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION, ... | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Sulfopin is a covalent inhibitor of Pin1 that blocks Myc-driven tumors in vivo.
Nat.Chem.Biol., 17, 2021
|
|
6M02
 
 | | cryo-EM structure of human Pannexin 1 channel | | Descriptor: | Pannexin-1 | | Authors: | Ronggui, Q, Lili, D, Jilin, Z, Xuekui, Y, Lei, W, Shujia, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-25 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of human heptameric Pannexin 1 channel.
Cell Res., 30, 2020
|
|
7DEG
 
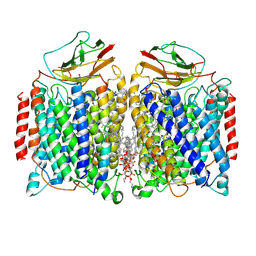 | | Cryo-EM structure of a heme-copper terminal oxidase dimer provides insights into its catalytic mechanism | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guoliang, Z, Shuangbo, Z. | | Deposit date: | 2020-11-04 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Unusual Homodimer of a Heme-Copper Terminal Oxidase Allows Itself to Utilize Two Electron Donors.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
