4WGZ
 
 | |
4WGY
 
 | |
2D1H
 
 | | Crystal structure of ST1889 protein from thermoacidophilic archaeon Sulfolobus tokodaii | | Descriptor: | 109aa long hypothetical transcriptional regulator | | Authors: | Shinkai, A, Sekine, S, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-22 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The putative DNA-binding protein Sto12a from the thermoacidophilic archaeon Sulfolobus tokodaii contains intrachain and interchain disulfide bonds.
J.Mol.Biol., 372, 2007
|
|
2DB0
 
 | | Crystal structure of PH0542 | | Descriptor: | 253aa long hypothetical protein | | Authors: | Nishino, A, Handa, N, Kishishita, S, Murayama, K, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PH0542
To be Published
|
|
2DBB
 
 | |
2DYQ
 
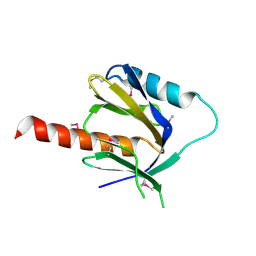 | | Crystal Structure of the C-terminal Phophotyrosine Interaction Domain of Human APBB3 | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 3 | | Authors: | Nishino, A, Saijo, S, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-16 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the C-terminal Phophotyrosine Interaction Domain of Human APBB3
To be Published
|
|
2E3V
 
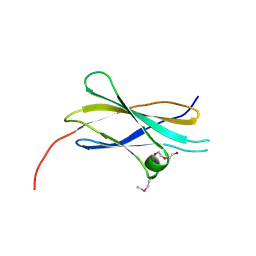 | | Crystal structure of the first fibronectin type III domain of neural cell adhesion molecule splicing isoform from human muscle culture lambda-4.4 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nishino, A, Saijo, S, Kishishita, S, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-11-30 | | Release date: | 2007-06-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the first fibronectin type III domain of neural cell adhesion molecule splicing isoform from human muscle culture lambda-4.4
To be Published
|
|
3U2H
 
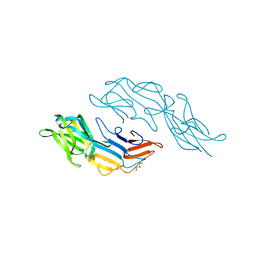 | | Crystal structure of the C-terminal DUF1608 domain of the Methanosarcina acetivorans S-layer (MA0829) protein | | Descriptor: | GLYCEROL, S-layer protein MA0829 | | Authors: | Chan, S, Phan, T, Ahn, C.J, Shin, A, Rohlin, L, Gunsalus, R.P, Arbing, M.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4U9P
 
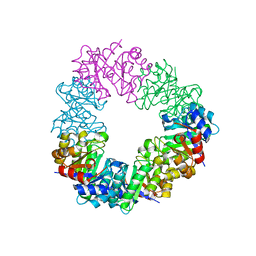 | | Structure of the methanofuran/methanopterin biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii | | Descriptor: | GLYCEROL, UPF0264 protein MJ1099 | | Authors: | Bobik, T.A, Morales, E, Shin, A, Cascio, D, Sawaya, M.R, Arbing, M, Rasche, M.E. | | Deposit date: | 2014-08-06 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the methanofuran/methanopterin-biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3MML
 
 | | Allophanate Hydrolase Complex from Mycobacterium smegmatis, Msmeg0435-Msmeg0436 | | Descriptor: | Allophanate hydrolase subunit 1, Allophanate hydrolase subunit 2, CHLORIDE ION | | Authors: | Kaufmann, M, Chernishof, I, Shin, A, Germano, D, Sawaya, M.R, Waldo, G.S, Arbing, M.A, Perry, J, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Allphanate Hydrolase Complex from M. smegmatis, Msmeg0435-Msmeg0436
To be Published
|
|
5DBN
 
 | | Crystal structure of AtoDA complex | | Descriptor: | Acetate CoA-transferase subunit alpha, Acetate CoA-transferase subunit beta, CHLORIDE ION, ... | | Authors: | Arbing, M.A, Koo, C.W, Shin, A, Medrano-Soto, A, Eisenberg, D. | | Deposit date: | 2015-08-21 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Crystal structure of AtoDA complex
To Be Published
|
|
5DB5
 
 | | Crystal structure of PLP-bound E. coli SufS (cysteine persulfide intermediate) in space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, CYSTEINE, ... | | Authors: | Arbing, M.A, Shin, A, Koo, C.W, Medrano-Soto, A, Eisenberg, D. | | Deposit date: | 2015-08-20 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of PLP-bound E. coli SufS (cysteine persulfide intermediate) in space group P21
To Be Published
|
|
5DUD
 
 | | Crystal structure of E. coli YbgJK | | Descriptor: | YbgJ, YbgK | | Authors: | Arbing, M.A, Kaufmann, M, Shin, A, Medrano-Soto, A, Cascio, D, Eisenberg, D. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of E. coli YbgJK
To Be Published
|
|
3Q4H
 
 | | Crystal structure of the Mycobacterium smegmatis EsxGH complex (MSMEG_0620-MSMEG_0621) | | Descriptor: | Low molecular weight protein antigen 7, Pe family protein | | Authors: | Chan, S, Harris, L, Kuo, E, Ahn, C, Zhou, T.T, Nguyen, L, Shin, A, Sawaya, M.R, Cascio, D, Arbing, M.A, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-12-23 | | Release date: | 2011-01-26 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
3U2G
 
 | | Crystal structure of the C-terminal DUF1608 domain of the Methanosarcina acetivorans S-layer (MA0829) protein | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Chan, S, Phan, T, Ahn, C.J, Shin, A, Rohlin, L, Gunsalus, R.P, Arbing, M.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5HJY
 
 | | Structure function studies of R. palustris RubisCO (I165T mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2016-01-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
5HJX
 
 | | Structure function studies of R. palustris RubisCO (A47V mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2016-01-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
5HQL
 
 | | Structure function studies of R. palustris RubisCO (A47V-M331A mutant; CABP-bound; no expression tag) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure function studies of R. palustris RubisCO
To Be Published
|
|
5HAO
 
 | | Structure function studies of R. palustris RubisCO (M331A mutant; CABP-bound) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Arbing, M.A, Shin, A, Satagopan, S, North, J.A, Tabita, F.R. | | Deposit date: | 2015-12-30 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
4OV0
 
 | | Structure of Bacteriorhdopsin Transferred from Amphipol A8-35 to a Lipidic Mesophase | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Polovinkin, V, Gushchin, I, Sintsov, M, Round, E, Balandin, T, Chervakov, P, Schevchenko, V, Utrobin, P, Popov, A, Borshchevskiy, V, Mishin, A, Kuklin, A, Willbold, D, Popot, J.L, Gordeliy, V. | | Deposit date: | 2014-02-19 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a membrane protein transferred from amphipol to a lipidic mesophase.
J.Membr.Biol., 247, 2014
|
|
8AMP
 
 | | Crystal structure of M.tuberculosis ferredoxin Fdx | | Descriptor: | FE (III) ION, FE3-S4 CLUSTER, Possible ferredoxin | | Authors: | Bukhdruker, S, Kavaleuski, A, Marin, E, Kapranov, I, Mishin, A, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
3BOY
 
 | | Crystal structure of the HutP antitermination complex bound to the HUT mRNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*GP*UP*UP*UP*UP*UP*AP*GP*UP*UP*UP*UP*UP*AP*GP*UP*UP*U)-3', HISTIDINE, Hut operon positive regulatory protein, ... | | Authors: | Kumarevel, T.S, Balasundaresan, D, Jeyakanthan, J, Shinkai, A, Yokoyama, S, Kumar, P.K.R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of HutP complexed with the 55-mer RNA
To be Published
|
|
2QQ4
 
 | | Crystal structure of Iron-sulfur cluster biosynthesis protein IscU (TTHA1736) from thermus thermophilus HB8 | | Descriptor: | Iron-sulfur cluster biosynthesis protein IscU, ZINC ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Agari, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-07-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Iron-sulfur cluster biosynthesis protein IscU (TTHA1736) from thermus thermophilus HB8
To be Published
|
|
2QQ1
 
 | | Crystal Structure Of Molybdenum Cofactor Biosynthesis (aq_061) Other Form From Aquifex Aeolicus Vf5 | | Descriptor: | Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Mahesh, S, Kanaujia, S.P, Ramakumar, S, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-07-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure Of Molybdenum Cofactor Biosynthesis (aq_061) Other Form From Aquifex Aeolicus Vf5
To be Published
|
|
2QYH
 
 | | Crystal structure of the hypothetical protein (gk1056) from geobacillus kaustophilus HTA426 | | Descriptor: | GLYCEROL, Hypothetical conserved protein, GK1056 | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-08-15 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the hypothetical protein (gk1056) from geobacillus kaustophilus HTA426
To be Published
|
|
