5ZXV
 
 | |
7CDI
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
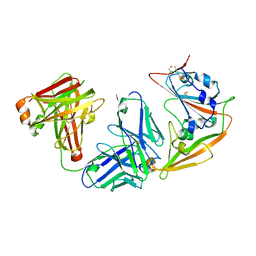
7CDJ
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1A3 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1A3 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.396 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
7TRL
 
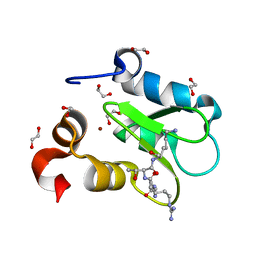 | | Crystal structure of human BIRC2 BIR3 domain in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, Histone H3, ... | | Authors: | Klein, B.J, Tencer, A.H, Kutateladze, T.G. | | Deposit date: | 2022-01-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Molecular basis for nuclear accumulation and targeting of the inhibitor of apoptosis BIRC2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TRM
 
 | | Crystal structure of human BIRC2 BIR3 domain in complex with inhibitor LCL-161 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, LCL-161, ... | | Authors: | Tencer, A.H, Klein, B.J, Kutateladze, T.G. | | Deposit date: | 2022-01-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for nuclear accumulation and targeting of the inhibitor of apoptosis BIRC2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4K94
 
 | | Crystal structure of KIT D4D5 fragment in complex with anti-Kit antibody Fab19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab19 heavy chain, Fab19 light chain, ... | | Authors: | Resheynyak, A.V, Boggon, T.J, Lax, I, Schlessinger, J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-10-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for KIT receptor tyrosine kinase inhibition by antibodies targeting the D4 membrane-proximal region.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K9E
 
 | | Crystal structure of KIT D4D5 fragment in complex with anti-Kit antibodies Fab79D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Mast/stem cell growth factor receptor Kit, heavy chain, ... | | Authors: | Resheynyak, A.V, Boggon, T.J, Lax, I, Schlessinger, J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-10-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for KIT receptor tyrosine kinase inhibition by antibodies targeting the D4 membrane-proximal region.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4N4G
 
 | |
4N4I
 
 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.3K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.3, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
8HGA
 
 | |
4N4H
 
 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.1K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.1, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
7WRP
 
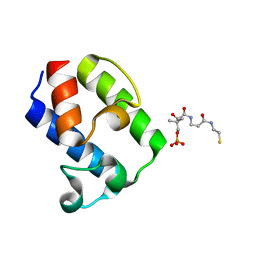 | | Crystal Structure of pks13-ACP domain from Corynebacterium diphtheriae | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Polyketide synthase involved in mycolic acid biosynthesis | | Authors: | Liu, X. | | Deposit date: | 2022-01-27 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Crystal structures of FadD32 and pks13-ACP domain from Corynebacterium diphtheriae.
Biochem.Biophys.Res.Commun., 590, 2022
|
|
5XTZ
 
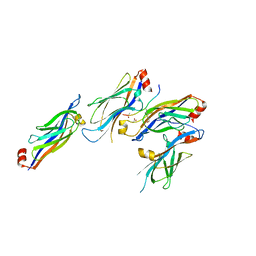 | | Crystal structure of GAS41 YEATS bound to H3K27ac peptide | | Descriptor: | ACETATE ION, THR-LYS-ALA-ALA-ARG-ALY-SER-ALA-PRO-ALA, YEATS domain-containing protein 4 | | Authors: | Li, H.T, Zhao, D. | | Deposit date: | 2017-06-21 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Gas41 links histone acetylation to H2A.Z deposition and maintenance of embryonic stem cell identity.
Cell Discov, 4, 2018
|
|
5T31
 
 | | Exploiting an Asp-Glu switch in Glycogen Synthase Kinase 3 to design paralog selective inhibitors for use in acute myeloid leukemia | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Stein, A.J, Holson, E.B, Wagner, F.F, Cambell, A.J. | | Deposit date: | 2016-08-24 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
7CKQ
 
 | |
7D7S
 
 | |
5Y3X
 
 | | Crystal structure of endo-1,4-beta-xylanase from Caldicellulosiruptor owensensis | | Descriptor: | Beta-xylanase | | Authors: | Liu, X, Sun, L.C, Zhang, Y.B, Liu, T.F, Xin, F.J. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Thermophilic Adaption Mechanism of Endo-1,4-beta-Xylanase from Caldicellulosiruptor owensensis.
J. Agric. Food Chem., 66, 2018
|
|
6A67
 
 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody FLD21.140 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLD21.140 Heavy Chain, FLD21.140 Light Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Complementary recognition of the receptor-binding site of highly pathogenic H5N1 influenza viruses by two human neutralizing antibodies.
J. Biol. Chem., 293, 2018
|
|
7KN3
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain complexed with a pre-pandemic antibody S-B8 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, S-B8 Fab heavy chain, ... | | Authors: | Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago.
Adv Sci, 9, 2022
|
|
6LSB
 
 | |
6LSD
 
 | |
6LS6
 
 | |
7X2M
 
 | | Crystal structure of nanobody 1-2C7 with SARS-CoV-2 RBD | | Descriptor: | 1-2C7, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2L
 
 | | Crystal structure of nanobody 3-2A2-4 with SARS-CoV-2 RBD | | Descriptor: | Nanobody 3-2A2-4, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2J
 
 | | Crystal structure of nanobody Nb70 with SARS-CoV RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nb70, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
