1WQR
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1998-02-09 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1XHB
 
 | | The Crystal Structure of UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase-T1 | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 1, ... | | Authors: | Fritz, T.A, Hurley, J.H, Trinh, L.B, Shiloach, J, Tabak, L.A. | | Deposit date: | 2004-09-17 | | Release date: | 2004-10-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The beginnings of mucin biosynthesis: The crystal structure of UDP-GalNAc:polypeptide {alpha}-N-acetylgalactosaminyltransferase-T1
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2ZQO
 
 | | Crystal structure of the earthworm R-type lectin C-half in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 29-kDa galactose-binding lectin, CADMIUM ION, ... | | Authors: | Suzuki, R, Kuno, A, Hasegawa, T, Hirabayashi, J, Kasai, K, Momma, M, Fujimoto, Z. | | Deposit date: | 2008-08-13 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sugar-complex structures of the C-half domain of the galactose-binding lectin EW29 from the earthworm Lumbricus terrestris
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZQN
 
 | | Crystal structure of the earthworm R-type lectin C-half in complex with Lactose | | Descriptor: | 29-kDa galactose-binding lectin, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Suzuki, R, Kuno, A, Hasegawa, T, Hirabayashi, J, Kasai, K, Momma, M, Fujimoto, Z. | | Deposit date: | 2008-08-13 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sugar-complex structures of the C-half domain of the galactose-binding lectin EW29 from the earthworm Lumbricus terrestris
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1ZZR
 
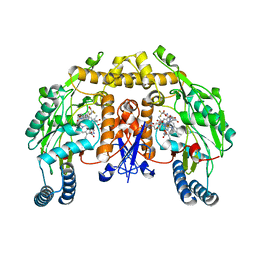 | | Rat nNOS D597N/M336V double mutant with L-N(omega)-Nitroarginine-(4R)-amino-L-proline amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, L-N(OMEGA)-NITROARGININE-(4R)-AMINO-L-PROLINE AMIDE, ... | | Authors: | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
1ZZT
 
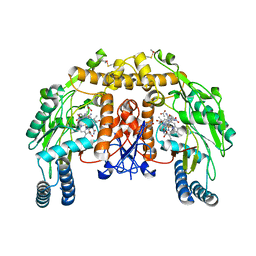 | | Bovine eNOS N368D/V106M double mutant with L-N(omega)-Nitroarginine-(4R)-Amino-L-Proline Amide Bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, L-N(OMEGA)-NITROARGININE-(4R)-AMINO-L-PROLINE AMIDE, ... | | Authors: | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-12-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
1ZZU
 
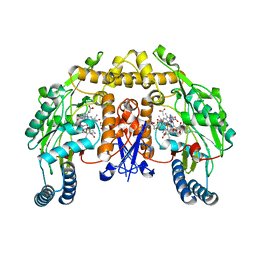 | | Rat nNOS D597N/M336V double mutant with L-N(omega)-Nitroarginine-2,4-L-Diaminobutyric Amide Bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
1ZZS
 
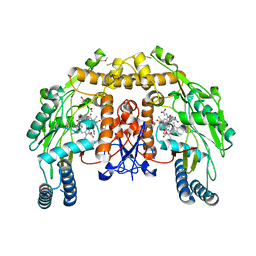 | | Bovine eNOS N368D single mutant with L-N(omega)-Nitroarginine-(4R)-Amino-L-Proline Amide Bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, GLYCEROL, ... | | Authors: | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
1ZZQ
 
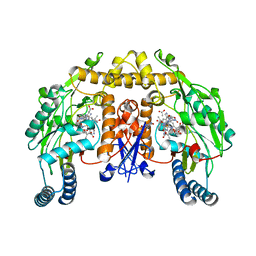 | | Rat nNOS D597N mutant with L-N(omega)-Nitroarginine-(4R)-amino-L-proline amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
3ALZ
 
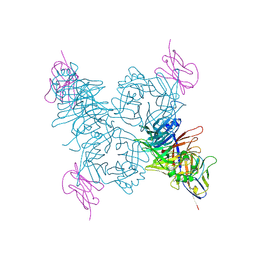 | | Crystal structure of the measles virus hemagglutinin bound to its cellular receptor SLAM (Form I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CDw150, Hemagglutinin | | Authors: | Hashiguchi, T, Ose, T, Kubota, M, Maita, N, Kamishikiryo, J, Maenaka, K, Yanagi, Y. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.515 Å) | | Cite: | Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM
Nat.Struct.Mol.Biol., 18, 2011
|
|
3ALX
 
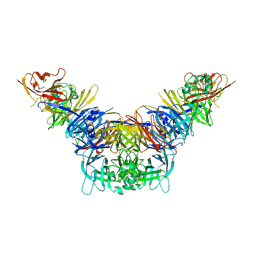 | | Crystal structure of the measles virus hemagglutinin bound to its cellular receptor SLAM (MV-H(L482R)-SLAM(N102H/R108Y) fusion) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin,LINKER,CDw150 | | Authors: | Hashiguchi, T, Ose, T, Kubota, M, Maita, N, Kamishikiryo, J, Maenaka, K, Yanagi, Y. | | Deposit date: | 2010-08-09 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM
Nat.Struct.Mol.Biol., 18, 2011
|
|
3ALW
 
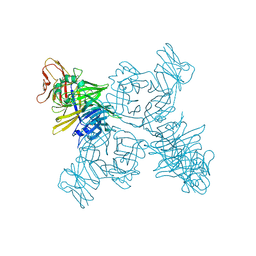 | | Crystal structure of the measles virus hemagglutinin bound to its cellular receptor SLAM (Form I, MV-H-SLAM(N102H/R108Y) fusion) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, CDw150 | | Authors: | Hashiguchi, T, Ose, T, Kubota, M, Maita, N, Kamishikiryo, J, Maenaka, K, Yanagi, Y. | | Deposit date: | 2010-08-09 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM
Nat.Struct.Mol.Biol., 18, 2011
|
|
2DM6
 
 | | Crystal structure of anti-configuration of indomethacin and leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complex | | Descriptor: | INDOMETHACIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent leukotriene B4 12-hydroxydehydrogenase, ... | | Authors: | Hori, T, Ishijima, J, Yokomizo, T, Ago, H, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-20 | | Release date: | 2006-11-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of anti-configuration of indomethacin and leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complex reveals the structural basis of broad spectrum indomethacin efficacy
J.Biochem.(Tokyo), 140, 2006
|
|
2A0Z
 
 | | The molecular structure of toll-like receptor 3 ligand binding domain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bell, J.K, Botos, I, Hall, P.R, Askins, J, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2005-06-17 | | Release date: | 2005-08-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular structure of the Toll-like receptor 3 ligand-binding domain
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2D5V
 
 | | Crystal structure of HNF-6alpha DNA-binding domain in complex with the TTR promoter | | Descriptor: | 5'-D(*AP*TP*TP*AP*TP*TP*GP*AP*CP*TP*TP*AP*GP*A)-3', 5'-D(*TP*CP*TP*AP*AP*GP*TP*CP*AP*AP*TP*AP*AP*T)-3', ACETATE ION, ... | | Authors: | Iyaguchi, D, Yao, M, Watanabe, N, Nishihira, J, Tanaka, I. | | Deposit date: | 2005-11-07 | | Release date: | 2006-12-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA recognition mechanism of the ONECUT homeodomain of transcription factor HNF-6
Structure, 15, 2007
|
|
3GJW
 
 | | PARP complexed with A968427 | | Descriptor: | 7-(pyrrolidin-1-ylmethyl)pyrrolo[1,2-a]quinoxalin-4(5H)-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Park, C.H. | | Deposit date: | 2009-03-09 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and SAR of novel tricyclic quinoxalinone inhibitors of poly(ADP-ribose)polymerase-1 (PARP-1)
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2QCY
 
 | |
3QDD
 
 | | HSP90A N-terminal domain in complex with BIIB021 | | Descriptor: | 6-chloro-9-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-9H-purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Arndt, J.W, Biamonte, M.A. | | Deposit date: | 2011-01-18 | | Release date: | 2012-07-18 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | EC144 Is a Potent Inhibitor of the Heat Shock Protein 90.
J.Med.Chem., 55, 2012
|
|
8HML
 
 | |
5GVU
 
 | |
7VWY
 
 | |
7W5Y
 
 | |
7W5W
 
 | |
2MJV
 
 | |
8JJE
 
 | | RBD of SARS-CoV2 spike protein with ACE2 decoy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kishikawa, J, Hirose, M, Kato, T, Okamoto, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An inhaled ACE2 decoy confers protection against SARS-CoV-2 infection in preclinical models.
Sci Transl Med, 15, 2023
|
|
