8F4L
 
 | |
7JU1
 
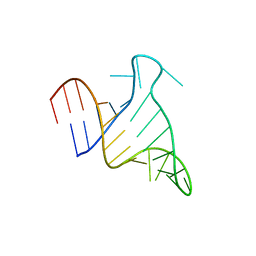 | | The FARFAR-NMR Ensemble of 29-mer HIV-1 Trans-activation Response Element RNA (N=20) | | Descriptor: | RNA (29-MER) | | Authors: | Shi, H, Rangadurai, A, Roy, R, Yesselman, J.D, Al-Hashimi, H.M. | | Deposit date: | 2020-08-18 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rapid and accurate determination of atomistic RNA dynamic ensemble models using NMR and structure prediction
Nat Commun, 11, 2020
|
|
6AI6
 
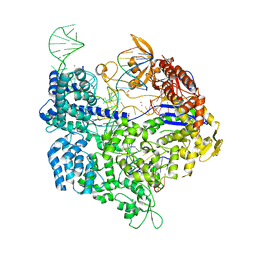 | | Crystal structure of SpCas9-NG | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, DNA (28-MER), ... | | Authors: | Nishimasu, H, Hirano, S, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-21 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineered CRISPR-Cas9 nuclease with expanded targeting space
Science, 361, 2018
|
|
8BQN
 
 | |
8EW2
 
 | |
8EW0
 
 | |
8F49
 
 | |
8F7Y
 
 | |
9C4C
 
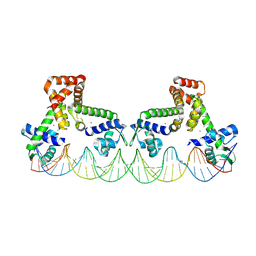 | | The structure of two MntR dimers bound to the native mnep promoter sequence | | Descriptor: | DNA (38-MER), DNA (39-MER), HTH-type transcriptional regulator MntR, ... | | Authors: | Shi, H, Fu, Y, Glasfeld, A, Ahuja, S. | | Deposit date: | 2024-06-04 | | Release date: | 2024-08-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for transcription activation through cooperative recruitment of MntR.
Nat Commun, 16, 2025
|
|
9C4D
 
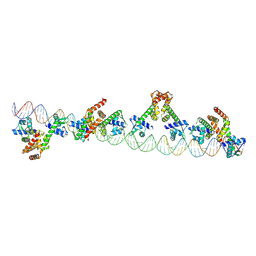 | | The structure of 4 MntR homodimers bound to the promoter sequence of mnep | | Descriptor: | DNA (77-MER), HTH-type transcriptional regulator MntR, MANGANESE (II) ION | | Authors: | Shi, H, Fu, Y, Glasfeld, A, Ahuja, S. | | Deposit date: | 2024-06-04 | | Release date: | 2024-08-14 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural basis for transcription activation through cooperative recruitment of MntR.
Nat Commun, 16, 2025
|
|
9C48
 
 | | Cryo-EM structure of the full-length human P2X4 receptor in the ATP-bound desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shi, H, Ditter, I.A, Oken, A.C, Mansoor, S.E. | | Deposit date: | 2024-06-03 | | Release date: | 2025-01-29 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Human P2X4 receptor gating is modulated by a stable cytoplasmic cap and a unique allosteric pocket.
Sci Adv, 11, 2025
|
|
9BQI
 
 | | Cryo-EM structure of BAY-1797 bound to the full-length human P2X4 receptor in the closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Shi, H, Ditter, I.A, Oken, A.C, Mansoor, S.E. | | Deposit date: | 2024-05-09 | | Release date: | 2025-01-29 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Human P2X4 receptor gating is modulated by a stable cytoplasmic cap and a unique allosteric pocket.
Sci Adv, 11, 2025
|
|
9BQH
 
 | | Cryo-EM structure of the full-length human P2X4 receptor in the apo closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Shi, H, Ditter, I.A, Oken, A.C, Mansoor, S.E. | | Deposit date: | 2024-05-09 | | Release date: | 2025-01-29 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Human P2X4 receptor gating is modulated by a stable cytoplasmic cap and a unique allosteric pocket.
Sci Adv, 11, 2025
|
|
3OPB
 
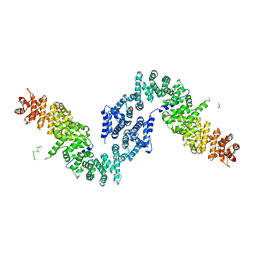 | | Crystal structure of She4p | | Descriptor: | SWI5-dependent HO expression protein 4 | | Authors: | Shi, H, Blobel, G. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | UNC-45/CRO1/She4p (UCS) protein forms elongated dimer and joins two myosin heads near their actin binding region.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4LL6
 
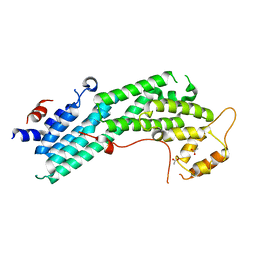 | | Structure of Myo4p globular tail domain. | | Descriptor: | ACETIC ACID, Myosin-4 | | Authors: | Shi, H, Singh, N, Esselborn, F, Blobel, G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a myosinbulletadaptor complex and pairing by cargo.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LL7
 
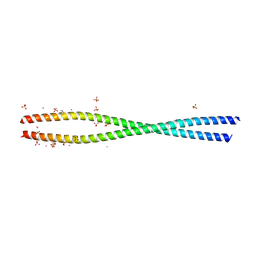 | | Structure of She3p amino terminus. | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DYSPROSIUM ION, ... | | Authors: | Shi, H, Singh, N, Esselborn, F, Blobel, G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of a myosinbulletadaptor complex and pairing by cargo.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LL8
 
 | | Complex of carboxy terminal domain of Myo4p and She3p middle fragment | | Descriptor: | Myosin-4, SWI5-dependent HO expression protein 3 | | Authors: | Shi, H, Singh, N, Esselborn, F, Blobel, G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.578 Å) | | Cite: | Structure of a myosinbulletadaptor complex and pairing by cargo.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5UBV
 
 | |
8HI2
 
 | |
8HHS
 
 | |
1OO0
 
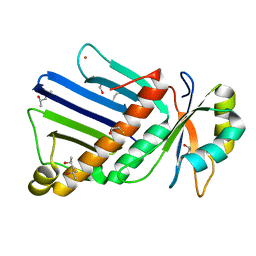 | |
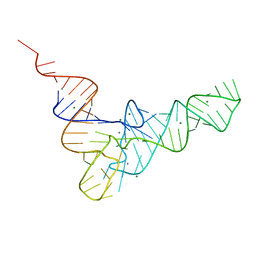
1EHZ
 
 | |
1HNS
 
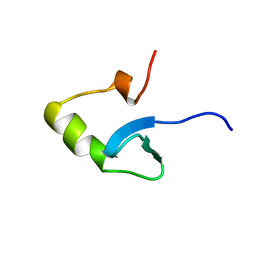 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
4YTR
 
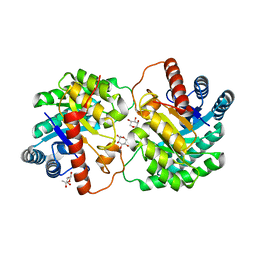 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy L-tagatose | | Descriptor: | 1-deoxy-L-tagatose, 1-deoxy-beta-L-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
4YTQ
 
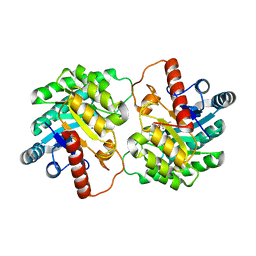 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy D-tagatose | | Descriptor: | 1-deoxy-D-tagatose, 1-deoxy-alpha-D-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
