5DAY
 
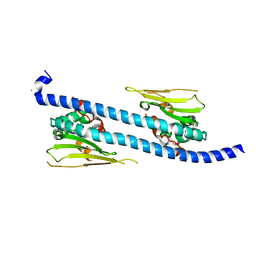 | | The structure of NAP1-Related Protein(NRP1) in Arabidopsis | | Descriptor: | CALCIUM ION, NAP1-related protein 1 | | Authors: | Zhu, Y, Rong, L, Yang, Y, Zhang, C, Feng, H.Y, Zheng, L.N, Shen, W.H, Ma, J.B, Dong, A.W. | | Deposit date: | 2015-08-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | The structure of NAP1-Related Protein(NRP1) in Arabidopsis
To Be Published
|
|
5XVW
 
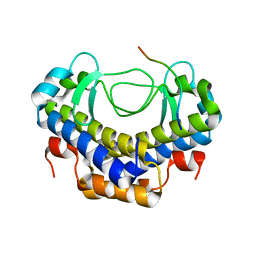 | |
5XVL
 
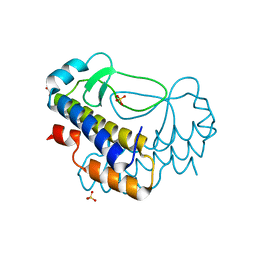 | | Crystal structure of AL2 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 2, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
7BP4
 
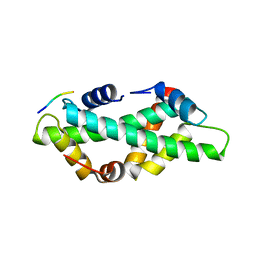 | |
7BP6
 
 | |
7BP2
 
 | |
7BP5
 
 | |
5Y53
 
 | |
5XVJ
 
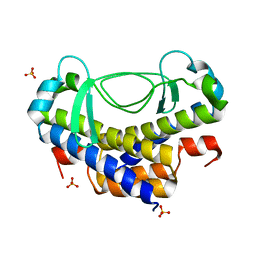 | | Crystal structure of AL7 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 7, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
5Y21
 
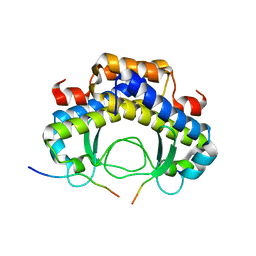 | |
6M2M
 
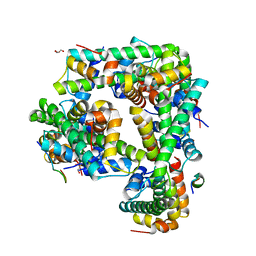 | |
6J0D
 
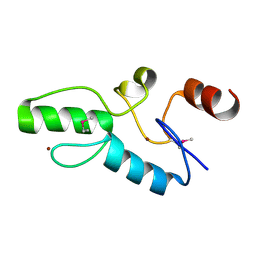 | | Crystal structure of OsSUF4 | | Descriptor: | ZINC ION, transcription factor | | Authors: | Wang, B, Luo, Q. | | Deposit date: | 2018-12-24 | | Release date: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The transcription factor OsSUF4 interacts with SDG725 in promoting H3K36me3 establishment.
Nat Commun, 10, 2019
|
|
7FDM
 
 | |
7FDL
 
 | |
7C7X
 
 | |
7CV9
 
 | | RNA methyltransferase METTL4 | | Descriptor: | GLYCEROL, Methyltransferase-like protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CVA
 
 | | RNA methyltransferase METTL4 | | Descriptor: | Methyltransferase-like protein 2 | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV6
 
 | | RNA methyltransferase METTL4 | | Descriptor: | 2'-O-methyladenosine 5'-(dihydrogen phosphate), Methyltransferase-like protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV8
 
 | | RNA methyltransferase METTL4 | | Descriptor: | GLYCEROL, Methyltransferase-like protein 2, SINEFUNGIN | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV7
 
 | | RNA methyltransferase METTL4 | | Descriptor: | Methyltransferase-like protein 2, S-ADENOSYLMETHIONINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7FDO
 
 | |
7FDN
 
 | |
5UIS
 
 | | Crystal structure of IRAK4 in complex with compound 12 | | Descriptor: | 4-{[(3R)-piperidin-3-yl]oxy}-6-[(propan-2-yl)oxy]quinoline-7-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIQ
 
 | | Crystal structure of IRAK4 in complex with compound 9 | | Descriptor: | 2-[(propan-2-yl)oxy]benzamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIR
 
 | | Crystal structure of IRAK4 in complex with compound 11 | | Descriptor: | 5-(4-cyanophenyl)-3-[(propan-2-yl)oxy]naphthalene-2-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
