8QXB
 
 | | TDP-43 amyloid fibrils: Morphology-2 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QXA
 
 | | TDP-43 amyloid fibrils: Morphology-1b | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QX9
 
 | | TDP-43 amyloid fibrils: Morphology-1a | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
7ZJ2
 
 | |
6H22
 
 | | Crystal structure of Mdm2 bound to a stapled peptide | | Descriptor: | 12-(dimethylamino)-3,10-diethyl-N,N,N-trimethyl-3,10-dihydrodibenzo[3,4:7,8]cycloocta[1,2-d:5,6-d']bis([1,2,3]triazole)-5-aminium, E3 ubiquitin-protein ligase Mdm2, Stapled peptide | | Authors: | Wang, X, Sharma, K, Spring, D.R, Hyvonen, M. | | Deposit date: | 2018-07-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Water-soluble, stable and azide-reactive strained dialkynes for biocompatible double strain-promoted click chemistry.
Org.Biomol.Chem., 17, 2019
|
|
4R6P
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6N
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8UAW
 
 | | Cryo-EM Structure of Brucella Abortus Lumazine Synthase (BLS) Engineered with Shiga Toxin II subunit B (Stx2B) | | Descriptor: | Shiga toxin II subunit B,6,7-dimethyl-8-ribityllumazine synthase 2 | | Authors: | Cristofalo, A.E, Sharma, A, Cerutti, M.L, Sharma, K, Zylberman, V, Goldbaum, F.A, Borgnia, M.J, Otero, L.H. | | Deposit date: | 2023-09-22 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Cryo-EM Structure of Brucella Abortus Lumazine Synthase (BLS) Engineered with Shiga Toxin II subunit B (Stx2B)
To Be Published
|
|
8UAV
 
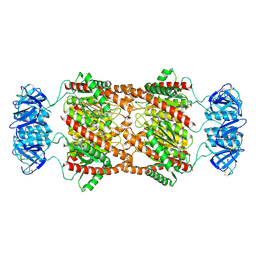 | | Cryo-EM Structure of Brucella Abortus Lumazine Synthase (BLS) Engineered with Shiga Toxin I subunit B (Stx1B) | | Descriptor: | Shiga toxin subunit B,6,7-dimethyl-8-ribityllumazine synthase 2 | | Authors: | Cristofalo, A.E, Sharma, A, Cerutti, M.L, Sharma, K, Zylberman, V, Goldbaum, F.A, Borgnia, M.J, Otero, L.H. | | Deposit date: | 2023-09-22 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Cryo-EM Structure of Brucella Abortus Lumazine Synthase (BLS) Engineered with Shiga Toxin I subunit B (Stx1B)
To Be Published
|
|
4R6R
 
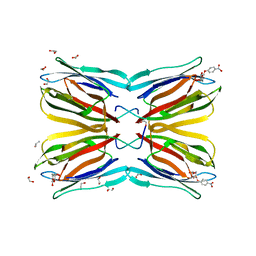 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrophenyl beta-D-galactopyranoside, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6Q
 
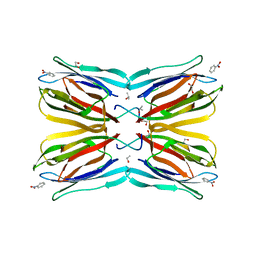 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6O
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
9CUO
 
 | | Crystal structure of CRBN with compound 3 | | Descriptor: | (3S)-3-(3-methyl-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-1-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, Protein cereblon, ... | | Authors: | Zheng, X, Ji, N, Campbell, V, Slavin, A, Zhu, X, Chen, D, Rong, H, Enerson, B, Mayo, M, Sharma, K, Browne, C.M, Klaus, C.R, Li, H, Massa, G, McDonald, A.A, Shi, Y, Sintchak, M, Skouras, S, Walther, D.M, Yuan, K, Zhang, Y, Kelleher, J, Guang, L, Luo, X, Mainolfi, N, Weiss, M.M. | | Deposit date: | 2024-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of KT-474─a Potent, Selective, and Orally Bioavailable IRAK4 Degrader for the Treatment of Autoimmune Diseases.
J.Med.Chem., 67, 2024
|
|
9B2A
 
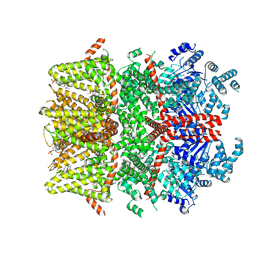 | | Cryo-EM structure of the mouse TRPM3 alpha 2 channel in complex with the neurosteroid pregnenolone sulfate and the synthetic agonist CIM 0216 | | Descriptor: | (2S)-2-(3,4-dihydroquinolin-1(2H)-yl)-N-(5-methyl-1,2-oxazol-3-yl)-2-phenylacetamide, Pregnenolone sulfate, Transient receptor potential cation channel, ... | | Authors: | Yin, Y, Park, C.G, Feng, S, Zhang, F, Guan, Z, Sharma, K, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2024-03-14 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Molecular basis of neurosteroid and anticonvulsant regulation of TRPM3.
Nat.Struct.Mol.Biol., 32, 2025
|
|
9B29
 
 | | Cryo-EM structure of the mouse TRPM3 alpha 2 channel in complex with cholesteryl hemisuccinate | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Yin, Y, Park, C.G, Feng, S, Zhang, F, Guan, Z, Sharma, K, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2024-03-14 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Molecular basis of neurosteroid and anticonvulsant regulation of TRPM3.
Nat.Struct.Mol.Biol., 32, 2025
|
|
9B28
 
 | | Cryo-EM structure of the mouse TRPM3 alpha 2 channel in complex with primidone | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel, subfamily M, ... | | Authors: | Yin, Y, Park, C.G, Feng, S, Zhang, F, Guan, Z, Sharma, K, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2024-03-14 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Molecular basis of neurosteroid and anticonvulsant regulation of TRPM3.
Nat.Struct.Mol.Biol., 32, 2025
|
|
4BRU
 
 | | Crystal structure of the yeast Dhh1-Edc3 complex | | Descriptor: | ATP-DEPENDENT RNA HELICASE DHH1, ENHANCER OF MRNA-DECAPPING PROTEIN 3 | | Authors: | Sharif, H, Ozgur, S, Sharma, K, Basquin, C, Urlaub, H, Conti, E. | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Structural Analysis of the Yeast Dhh1-Pat1 Complex Reveals How Dhh1 Engages Pat1, Edc3 and RNA in Mutually Exclusive Interactions
Nucleic Acids Res., 41, 2013
|
|
4BRW
 
 | | Crystal structure of the yeast Dhh1-Pat1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-DEPENDENT RNA HELICASE DHH1, DNA TOPOISOMERASE 2-ASSOCIATED PROTEIN PAT1, ... | | Authors: | Sharif, H, Ozgur, S, Sharma, K, Basquin, C, Urlaub, H, Conti, E. | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Structural Analysis of the Yeast Dhh1-Pat1 Complex Reveals How Dhh1 Engages Pat1, Edc3 and RNA in Mutually Exclusive Interactions
Nucleic Acids Res., 41, 2013
|
|
8R4A
 
 | |
5J50
 
 | | Structure of tetrameric jacalin complexed with Gal beta-(1,3) GalNAc-alpha-OPNP | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Surolia, A, Vijayan, M. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of tetrameric jacalin complexed with Gal beta-(1,3) GalNAc-alpha-OPNP
To Be Published
|
|
5J4T
 
 | | Structure of tetrameric jacalin complexed with GlcNAc beta-(1,3) Gal-beta-OMe | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-methyl beta-D-galactopyranoside, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Surolia, A, Vijayan, M. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Distortion of the ligand molecule as a strategy for modulating binding affinity: Further studies involving complexes of jacalin with beta-substituted disaccharides.
IUBMB Life, 69, 2017
|
|
5J4X
 
 | | Structure of tetrameric jacalin complexed with Gal beta-(1,3) Gal-beta-OMe | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Surolia, A, Vijayan, M. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Distortion of the ligand molecule as a strategy for modulating binding affinity: Further studies involving complexes of jacalin with beta-substituted disaccharides.
IUBMB Life, 69, 2017
|
|
5JM1
 
 | | Structure of tetrameric jacalin complexed with a trisaccharide, Gal alpha-(1,3) Gal beta-(1,4) Gal | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Surolia, A, Vijayan, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Effect of linkage on the location of reducing and nonreducing sugars bound to jacalin.
IUBMB Life, 68, 2016
|
|
5J51
 
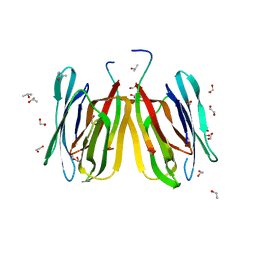 | | Structure of tetrameric jacalin complexed with Gal alpha-(1,4) Gal | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Surolia, A, Vijayan, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Effect of linkage on the location of reducing and nonreducing sugars bound to jacalin.
IUBMB Life, 68, 2016
|
|
8V2F
 
 | | Crystal structure of IRAK4 kinase domain with compound 9 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
