1HA1
 
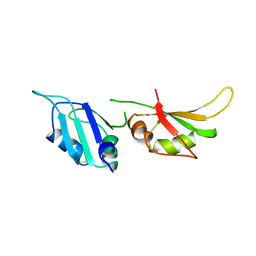 | | HNRNP A1 (RBD1,2) FROM HOMO SAPIENS | | Descriptor: | HNRNP A1 | | Authors: | Shamoo, Y, Krueger, U, Rice, L, Williams, K.R, Steitz, T.A. | | Deposit date: | 1996-10-30 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 A resolution.
Nat.Struct.Biol., 4, 1997
|
|
1GPC
 
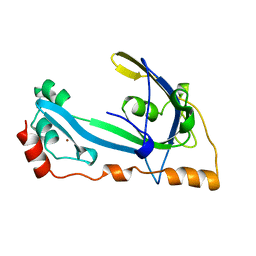 | | CORE GP32, DNA-BINDING PROTEIN | | Descriptor: | PROTEIN (CORE GP32), ZINC ION | | Authors: | Shamoo, Y, Friedman, A.M, Parsons, M.R, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1995-06-01 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA.
Nature, 376, 1995
|
|
6C0Z
 
 | |
6C84
 
 | |
6BWS
 
 | |
1B77
 
 | |
1B8H
 
 | | SLIDING CLAMP, DNA POLYMERASE | | Descriptor: | DNA POLYMERASE PROCESSIVITY COMPONENT, DNA POLYMERASE fragment | | Authors: | Shamoo, Y, Steitz, T.A. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CLQ
 
 | |
8D84
 
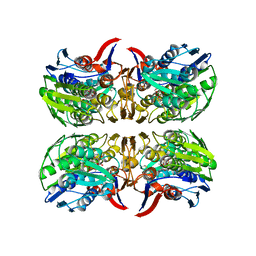 | | E. faecium MurAA in complex with UDP-N-acetylmuramic acid (UNAM) and a covalent adduct of PEP with Cys119 | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Zhou, Y, Shamoo, Y. | | Deposit date: | 2022-06-07 | | Release date: | 2023-07-05 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Enolpyruvate transferase MurAA A149E , identified during adaptation of Enterococcus faecium to daptomycin, increases stability of MurAA-MurG interaction.
J.Biol.Chem., 299, 2023
|
|
8U55
 
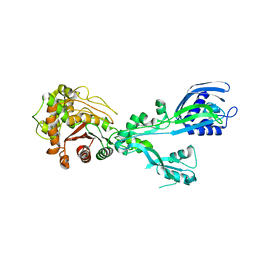 | |
7TB0
 
 | | E. faecium MurAA in complex with fosfomycin and UNAG | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SODIUM ION, ... | | Authors: | Zhou, Y, Shamoo, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enolpyruvate transferase MurAA A149E , identified during adaptation of Enterococcus faecium to daptomycin, increases stability of MurAA-MurG interaction.
J.Biol.Chem., 299, 2023
|
|
5HEV
 
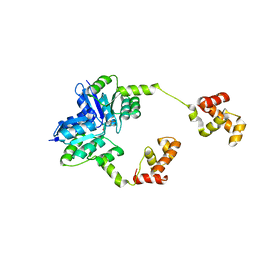 | |
3P9U
 
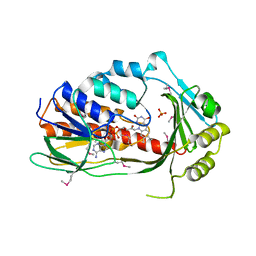 | | Crystal structure of TetX2 from Bacteroides thetaiotaomicron with substrate analogue | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TetX2 protein | | Authors: | Walkiewicz, K, Davlieva, M, Sun, C, Lau, K, Shamoo, Y. | | Deposit date: | 2010-10-18 | | Release date: | 2011-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of Bacteroides thetaiotaomicron TetX2: a tetracycline degrading monooxygenase at 2.8 A resolution.
Proteins, 79, 2011
|
|
4PDB
 
 | |
2A1K
 
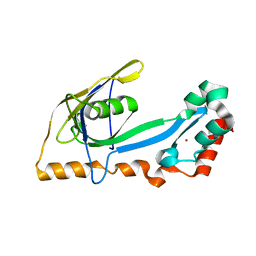 | | RB69 single-stranded DNA binding protein core domain | | Descriptor: | ZINC ION, gp32 single stranded DNA binding protein | | Authors: | Sun, S, Geng, L, Shamoo, Y. | | Deposit date: | 2005-06-20 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and enzymatic properties of a chimeric bacteriophage RB69 DNA polymerase and single-stranded DNA binding protein with increased processivity.
Proteins, 65, 2006
|
|
1PO6
 
 | |
1PGZ
 
 | |
4WUH
 
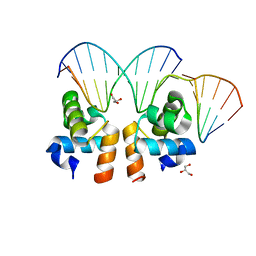 | | Crystal structure of E. faecalis DNA binding domain LiaR wild type complexed with 22bp DNA | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*CP*G)-3'), DNA (5'-D(P*GP*GP*AP*CP*TP*TP*AP*AP*GP*AP*AP*CP*GP*AP*TP*TP*T)-3'), DNA (5'-D(P*TP*TP*CP*TP*TP*AP*AP*GP*TP*CP*C)-3'), ... | | Authors: | Davlieva, M, Shamoo, Y. | | Deposit date: | 2014-10-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin.
Nucleic Acids Res., 43, 2015
|
|
4WSZ
 
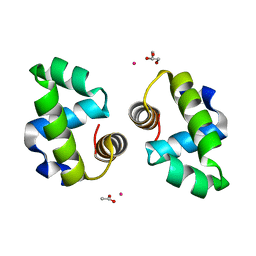 | |
4WT0
 
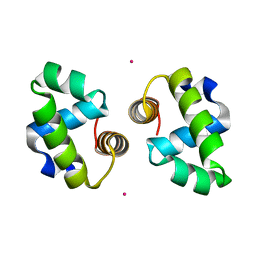 | |
4WUL
 
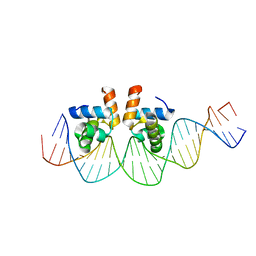 | |
4WU4
 
 | | Crystal structure of E. faecalis DNA binding domain LiaRD191N complexed with 22bp DNA | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*CP*GP*TP*TP*CP*TP*TP*AP*AP*GP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*CP*TP*TP*AP*AP*GP*AP*AP*CP*GP*AP*TP*TP*T)-3'), GLYCEROL, ... | | Authors: | Davlieva, M, Shamoo, Y. | | Deposit date: | 2014-10-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin.
Nucleic Acids Res., 43, 2015
|
|
3V3O
 
 | |
3V3N
 
 | | Crystal structure of TetX2 T280A: an adaptive mutant in complex with minocycline | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Walkiewicz, K, Shamoo, Y. | | Deposit date: | 2011-12-13 | | Release date: | 2013-01-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of TetX2 T280A: an adaptive mutant in complex with minocycline
To be Published
|
|
2BCX
 
 | | Crystal structure of calmodulin in complex with a ryanodine receptor peptide | | Descriptor: | CALCIUM ION, Calmodulin, Ryanodine receptor 1 | | Authors: | Maximciuc, A.A, Shamoo, Y, MacKenzie, K.R. | | Deposit date: | 2005-10-19 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex of calmodulin with a ryanodine receptor target reveals a novel, flexible binding mode.
Structure, 14, 2006
|
|
