8BH1
 
 | | Core divisome complex FtsWIQBL from Pseudomonas aeruginosa | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | Authors: | Kaeshammer, L, van den Ent, F, Jeffery, M, Lowe, J. | | Deposit date: | 2022-10-28 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the bacterial divisome core complex and antibiotic target FtsWIQBL.
Nat Microbiol, 8, 2023
|
|
1YR2
 
 | |
1C28
 
 | |
6S85
 
 | | Cutting state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (31-MER), DNA (32-MER), ... | | Authors: | Kaeshammer, L, Saathoff, J.H, Gut, F, Bartho, J, Alt, A, Kessler, B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-07-08 | | Release date: | 2019-09-04 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex.
Mol.Cell, 76, 2019
|
|
6S6V
 
 | | Resting state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ATPgS | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Nuclease SbcCD subunit C, ... | | Authors: | Kaeshammer, L, Saathoff, J.H, Gut, F, Bartho, J, Alt, A, Kessler, B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-07-03 | | Release date: | 2019-09-04 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex.
Mol.Cell, 76, 2019
|
|
1NCI
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, URANYL (VI) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NEU
 
 | | STRUCTURE OF MYELIN MEMBRANE ADHESION MOLECULE P0 | | Descriptor: | MYELIN P0 PROTEIN | | Authors: | Shapiro, L, Doyle, J.P, Hensley, P, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1996-09-24 | | Release date: | 1997-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin.
Neuron, 17, 1996
|
|
1NCH
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NCG
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
6AUL
 
 | | Artificial Metalloproteins Containing a Co4O4 Active Site - 2xm-S112Y-b | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Olshansky, L, Vallapurakal, J, Huerta-Lavorie, R, Nguyen, A.I, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
6AUH
 
 | | Artificial Metalloproteins Containing a Co4O4 Active Site - 2xm-S112Y-a | | Descriptor: | N-biotin-C-Co4(mu3-O)4(OAc)(Py)3(H2O)3-beta-alanine, Streptavidin | | Authors: | Olshansky, L, Vallapurakal, J, Huerta-Lavorie, R, Nguyen, A.I, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
6AUC
 
 | | Artificial metalloproteins containing a Co4O4 active site - 2xm-Sav | | Descriptor: | N-biotin-C-Co4(mu3-O)4(Py)4(H2O)4-beta-alanine, Streptavidin | | Authors: | Olshansky, L, Vallapurackal, J, Huerta-Lavorie, R, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
6AUO
 
 | | Artificial Metalloproteins Containing a Co4O4 Active Site - 2xm-S112F | | Descriptor: | N-biotin-C-Co4(mu3-O)4(Py)3(H2O)4-beta-alanine, Streptavidin | | Authors: | Olshansky, L, Vallapurakal, J, Huerta-Lavorie, R, Nguyen, A.I, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
6AUE
 
 | | Artificial Metalloproteins Containing a Co4O4 Active Site - 2xm-S112Y-b | | Descriptor: | N-biotin-C-Co4(mu3-O)4(OAc)(Py)4(H2O)3-beta-alanine, Streptavidin | | Authors: | Olshansky, L, Vallapurakal, J, Huerta-Lavorie, R, Nguyen, A.I, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
2PQE
 
 | | Solution structure of proline-free mutant of staphylococcal nuclease | | Descriptor: | Thermonuclease | | Authors: | Shan, L, Tong, Y, Xie, T, Wang, M, Wang, J. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Restricted backbone conformational and motional flexibilities of loops containing peptidyl-proline bonds dominate the enzyme activity of staphylococcal nuclease.
Biochemistry, 46, 2007
|
|
7W2I
 
 | | Crystal structure of LOG (Rv1205) from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Cytokinin riboside 5'-monophosphate phosphoribohydrolase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shang, L, Zhang, G. | | Deposit date: | 2021-11-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cytokinin-producing enzyme "lonely guy" (LOG) from Mycobacterium tuberculosis.
Biochem.Biophys.Res.Commun., 598, 2022
|
|
1U9U
 
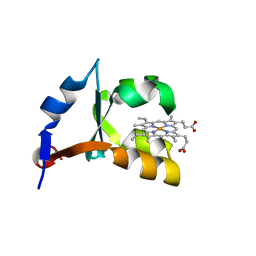 | | Crystal structure of F58Y mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U9M
 
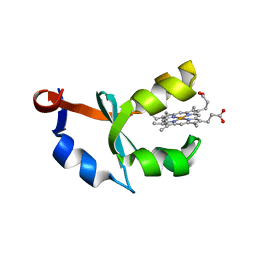 | | Crystal structure of F58W mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-10 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7U5V
 
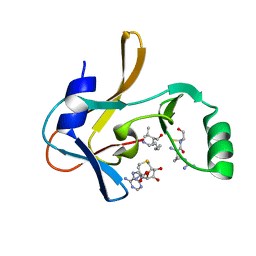 | | Crystal structure of the Mixed Lineage Leukaemia (MLL1) SET Domain with the cofactor product S-Adenosylhomocysteine and Borealin peptide | | Descriptor: | Borealin, Histone-lysine N-methyltransferase 2A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | An, S, Cho, U.S, Oh, H, Sha, L, Xu, J, Kim, S, Yang, W, An, W, Dou, Y. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Non-canonical MLL1 activity regulates centromeric phase separation and genome stability.
Nat.Cell Biol., 25, 2023
|
|
1C3H
 
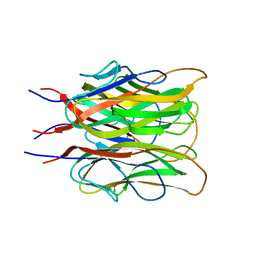 | | ACRP30 CALCIUM COMPLEX | | Descriptor: | 30 KD ADIPOCYTE COMPLEMENT-RELATED PROTEIN PRECURSOR, CALCIUM ION | | Authors: | Shapiro, L, Boggon, T, Scherer, P. | | Deposit date: | 1999-07-27 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ACRP30 calcium complex
To Be Published
|
|
2BKL
 
 | | Structural and Mechanistic Analysis of Two Prolyl Endopeptidases: Role of Inter-Domain Dynamics in Catalysis and Specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, N-[(BENZYLOXY)CARBONYL]-L-ALANYL-L-PROLINE, PROLYL ENDOPEPTIDASE, ... | | Authors: | Khosla, C, Shan, L, Mathews, I.I. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Mechanistic Analysis of Two Prolyl Endopeptidases: Role of Interdomain Dynamics in Catalysis and Specificity
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6WUD
 
 | |
6WU7
 
 | |
6WIB
 
 | | Next generation monomeric IgG4 Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant gamma 4, ZINC ION | | Authors: | Oganesyan, V.Y, Shan, L, Dall'Acqua, W, van Dyk, N. | | Deposit date: | 2020-04-09 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | In vivo pharmacokinetic enhancement of monomeric Fc and monovalent bispecific designs through structural guidance.
Commun Biol, 4, 2021
|
|
6WMH
 
 | |
