5NZ5
 
 | |
5NYC
 
 | |
5NY7
 
 | |
5NXZ
 
 | |
2DPP
 
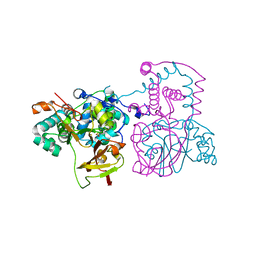 | | Crystal structure of thermostable Bacillus sp. RAPc8 nitrile hydratase | | Descriptor: | COBALT (II) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit | | Authors: | Tsekoa, T.L, Tastan-Bishop, A.O, Cameron, R.A, Sewell, B.T, Sayed, M.F, Cowan, D.A. | | Deposit date: | 2006-05-12 | | Release date: | 2007-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of thermostable Bacillus sp. RAPc8 nitrile hydratse
To be Published
|
|
4KZF
 
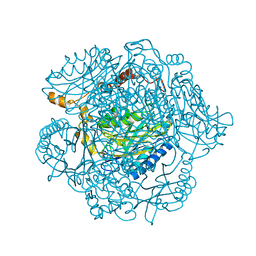 | | The mechanism of the amidases: The effect of the mutation E142L in the amidase from Geobacillus pallidus | | Descriptor: | Aliphatic amidase, CHLORIDE ION | | Authors: | Weber, B.W, Sewell, B.T, Kimani, S.W, Varsani, A, Cowan, D.A, Hunter, R. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The mechanism of the amidases: mutating the glutamate adjacent to the catalytic triad inactivates the enzyme due to substrate mispositioning.
J.Biol.Chem., 288, 2013
|
|
3HGJ
 
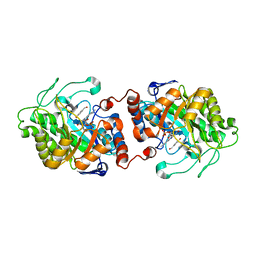 | | Old Yellow Enzyme from Thermus scotoductus SA-01 complexed with p-hydroxy-benzaldehyde | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZALDEHYDE | | Authors: | Opperman, D.J, Sewell, B.T, Litthauer, D, Isupov, M.N, Littlechild, J.A, van Heerden, E. | | Deposit date: | 2009-05-14 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01
Biochem.Biophys.Res.Commun., 393, 2010
|
|
3HHT
 
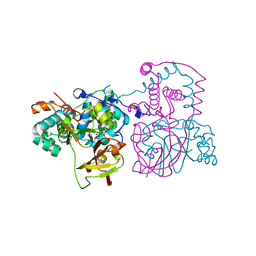 | | A mutant of the nitrile hydratase from Geobacillus pallidus having enhanced thermostability | | Descriptor: | CHLORIDE ION, COBALT (III) ION, Nitrile hydratase alpha subunit, ... | | Authors: | Van Wyk, J.C, Sewell, B.T, Cowan, D.A, Sayed, M.F, Tsekoa, T.L, Tastan Bishop, A.O. | | Deposit date: | 2009-05-17 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | The high-resolution structure of a Cobalt containing nitrile hydratase
To be published
|
|
3HF3
 
 | | Old Yellow Enzyme from Thermus scotoductus SA-01 | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE, SULFATE ION | | Authors: | Opperman, D.J, Sewell, B.T, Litthauer, D, Isupov, M.N, Littlechild, J.A, van Heerden, E. | | Deposit date: | 2009-05-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01
Biochem.Biophys.Res.Commun., 393, 2010
|
|
3L3N
 
 | | Testis ACE co-crystal structure with novel inhibitor lisW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, CHLORIDE ION, ... | | Authors: | Watermeyer, J.M, Kroger, W.L, O'Neil, H.G, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2009-12-17 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of domain-selective inhibitor binding in angiotensin-converting enzyme using a novel derivative of lisinopril.
Biochem.J., 428, 2010
|
|
6T1T
 
 | |
4GYN
 
 | | The E142L mutant of the amidase from Geobacillus pallidus | | Descriptor: | Aliphatic amidase, CHLORIDE ION | | Authors: | Weber, B.W, Sewell, B.T, Kimani, S.W, Varsani, A, Cowan, D.A, Hunter, R. | | Deposit date: | 2012-09-05 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The mechanism of the amidases: mutating the glutamate adjacent to the catalytic triad inactivates the enzyme due to substrate mispositioning.
J.Biol.Chem., 288, 2013
|
|
4GYL
 
 | | The E142L mutant of the amidase from Geobacillus pallidus showing the result of Michael addition of acrylamide at the active site cysteine | | Descriptor: | Aliphatic amidase, CHLORIDE ION, PROPIONAMIDE | | Authors: | Weber, B.W, Sewell, B.T, Kimani, S.W, Varsani, A, Cowan, D.A, Hunter, R. | | Deposit date: | 2012-09-05 | | Release date: | 2013-08-21 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The mechanism of the amidases: mutating the glutamate adjacent to the catalytic triad inactivates the enzyme due to substrate mispositioning.
J.Biol.Chem., 288, 2013
|
|
4IZT
 
 | |
4IZU
 
 | |
4IZV
 
 | |
4IZW
 
 | |
4IZS
 
 | |
7OVG
 
 | |
7Q49
 
 | | Local refinement structure of the N-domain of full-length, monomeric, soluble somatic angiotensin I-converting enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ZINC ION, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-29 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
7Q3Y
 
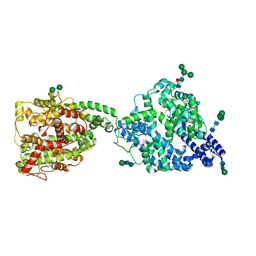 | | Structure of full-length, monomeric, soluble somatic angiotensin I-converting enzyme showing the N- and C-terminal ellipsoid domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-29 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
7Q4C
 
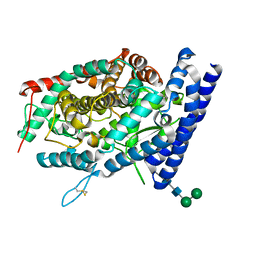 | | Local refinement structure of the C-domain of full-length, monomeric, soluble somatic angiotensin I-converting enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-30 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
7Q4E
 
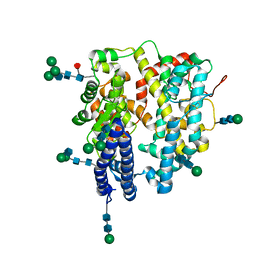 | | Local refinement structure of a single N-domain of full-length, dimeric, soluble somatic angiotensin I-converting enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-30 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
7Q4D
 
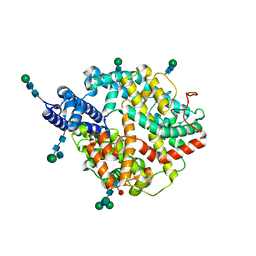 | | Local refinement structure of the two interacting N-domains of full-length, dimeric, soluble somatic angiotensin I-converting enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-30 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
6FKX
 
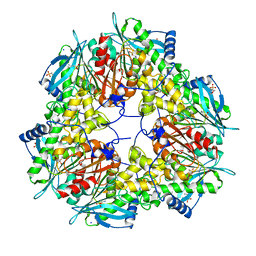 | | Crystal structure of an acetyl xylan esterase from a desert metagenome | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetyl xylan esterase, FORMIC ACID, ... | | Authors: | Adesioye, F.A, Makhalanyane, T.P, Vikram, S, Sewell, B.T, Schubert, W, Cowan, D.A. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Characterization and Directed Evolution of a Novel Acetyl Xylan Esterase Reveals Thermostability Determinants of the Carbohydrate Esterase 7 Family.
Appl. Environ. Microbiol., 84, 2018
|
|
