6ZCT
 
 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | ZINC ION, nsp10 | | Authors: | Rogstam, A, Nyblom, M, Christensen, S, Sele, C, Lindvall, T, Rasmussen, A.A, Andre, I, Fisher, S.Z, Knecht, W, Kozielski, F. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
7ORW
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00265 | | Descriptor: | 1H-benzimidazol-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORU
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00221 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORV
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
6F2O
 
 | |
9FWP
 
 | | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00198 | | Descriptor: | Guanine-N7 methyltransferase nsp14, MAGNESIUM ION, N-methylbenzamide, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00198
To Be Published
|
|
9FWH
 
 | | Crystal Structure of SARS-CoV-2 NSP10-ExoN in complex with VT00019 | | Descriptor: | (4R)-4-phenyl-1,2-thiazolidine 1,1-dioxide, Guanine-N7 methyltransferase nsp14, Non-structural protein 10, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of SARS-CoV-2 NSP10-ExoN in complex with VT00019
To Be Published
|
|
9FWQ
 
 | | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00218 | | Descriptor: | 5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyridine, Guanine-N7 methyltransferase nsp14, MAGNESIUM ION, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00218
To Be Published
|
|
9FWU
 
 | | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00421 | | Descriptor: | DIMETHYL SULFOXIDE, Guanine-N7 methyltransferase nsp14, N,N-dimethyl-3-oxidanyl-benzamide, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.425 Å) | | Cite: | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00421
To Be Published
|
|
9FWO
 
 | | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00216 | | Descriptor: | 1-methylpyrrole-2-carboxamide, Guanine-N7 methyltransferase nsp14, MAGNESIUM ION, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.179 Å) | | Cite: | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00216
To Be Published
|
|
9FWL
 
 | | Ensemble model of ligand-free SARS-CoV-2 NSP10-NSP14 (ExoN) and in complex with partially bound VT00025 | | Descriptor: | 3-phenylthiophene-2-carboxamide, DIMETHYL SULFOXIDE, Guanine-N7 methyltransferase nsp14, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Ensemble model of ligand-free SARS-CoV-2 NSP10-NSP14 (ExoN) and in complex with partially bound VT00025
To Be Published
|
|
9FWJ
 
 | | Ensemble model of ligand-free SARS-CoV-2 NSP10-NSP14 (ExoN) and in complex with partially bound VT00079 | | Descriptor: | 2-methoxybenzamide, Guanine-N7 methyltransferase nsp14, MAGNESIUM ION, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Ensemble model of ligand-free SARS-CoV-2 NSP10-NSP14 (ExoN) and in complex with partially bound VT00079
To Be Published
|
|
9FWN
 
 | | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00219 | | Descriptor: | 1-methyl-1-(phenylmethyl)urea, DIMETHYL SULFOXIDE, Guanine-N7 methyltransferase nsp14, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00219
To Be Published
|
|
9FWM
 
 | | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00180 | | Descriptor: | 1H-indole-3-carboxamide, DIMETHYL SULFOXIDE, Guanine-N7 methyltransferase nsp14, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.574 Å) | | Cite: | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00180
To Be Published
|
|
9FWS
 
 | | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00258 | | Descriptor: | 7-METHOXY-1H-INDAZOLE, DIMETHYL SULFOXIDE, Guanine-N7 methyltransferase nsp14, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.433 Å) | | Cite: | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00258
To Be Published
|
|
9FWT
 
 | | Ensemble model of ligand-free SARS-CoV-2 NSP10-NSP14 (ExoN) and in complex with partially bound VT00259 | | Descriptor: | DIMETHYL SULFOXIDE, Guanine-N7 methyltransferase nsp14, Non-structural protein 10, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Ensemble model of ligand-free SARS-CoV-2 NSP10-NSP14 (ExoN) and in complex with partially bound VT00259
To Be Published
|
|
9FWR
 
 | | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00249 | | Descriptor: | (4R)-4-phenyl-1,3-oxazolidin-2-one, Guanine-N7 methyltransferase nsp14, MAGNESIUM ION, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Crystal Structure of SARS-CoV-2 NSP10-NSP14 (ExoN) in complex with VT00249
To Be Published
|
|
9FWI
 
 | | Ensemble model of ligand-free SARS-CoV-2 NSP10-NSP14 (ExoN) and in complex with partially bound VT00025 | | Descriptor: | (3-oxidanylazetidin-1-yl)-phenyl-methanone, DIMETHYL SULFOXIDE, Guanine-N7 methyltransferase nsp14, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.533 Å) | | Cite: | Ensemble model of ligand-free SARS-CoV-2 NSP10-NSP14 (ExoN) and in complex with partially bound VT00025
To Be Published
|
|
5A5C
 
 | | Structure of an engineered neuronal LRRTM2 adhesion molecule | | Descriptor: | LRRTM | | Authors: | Paatero, A, Rosti, K, Shkumatov, A.V, Brunello, C, Kysenius, K, Huttunen, H, Kajander, T. | | Deposit date: | 2015-06-17 | | Release date: | 2016-02-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal Structure of an Engineered Lrrtm2 Synaptic Adhesion Molecule and a Model for Neurexin Binding.
Biochemistry, 55, 2016
|
|
6ZPE
 
 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Replicase polyprotein 1ab, ... | | Authors: | Fisher, S.Z, Kozielski, F. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
5K4Z
 
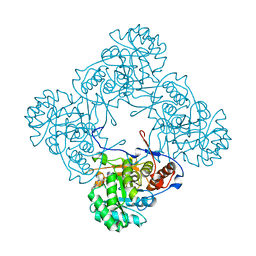 | | M. thermoresistible IMPDH in complex with IMP and Compound 6 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(4-fluorophenyl)-4-(2~{H}-indazol-6-ylsulfamoyl)-3,5-dimethyl-1~{H}-pyrrole-2-carboxamide | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-05-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
5K4X
 
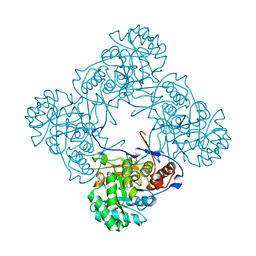 | | M. thermoresistible IMPDH in complex with IMP and Compound 1 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(2~{H}-indazol-6-yl)-3,5-dimethyl-1~{H}-pyrazole-4-sulfonamide | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-05-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
4IR4
 
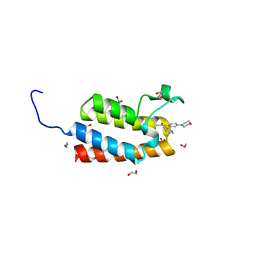 | | Crystal Structure of the bromodomain of human BAZ2B in complex with 1-[7-(morpholin-4-yl)-1-(pyridin-2-yl)indolizin-3-yl]ethanone (GSK2834113A) | | Descriptor: | 1,2-ETHANEDIOL, 1-[7-(morpholin-4-yl)-1-(pyridin-2-yl)indolizin-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
4IR5
 
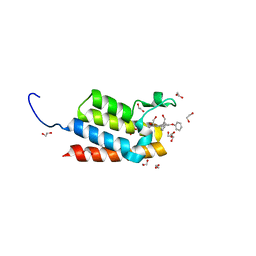 | | Crystal Structure of the bromodomain of human BAZ2B in complex with 1-{1-[2-(hydroxymethyl)phenyl]-7-phenoxyindolizin-3-yl}ethanone (GSK2847449A) | | Descriptor: | 1,2-ETHANEDIOL, 1-{1-[2-(hydroxymethyl)phenyl]-7-phenoxyindolizin-3-yl}ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
