4A11
 
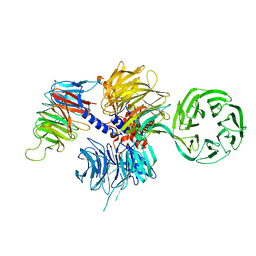 | | Structure of the hsDDB1-hsCSA complex | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, DNA EXCISION REPAIR PROTEIN ERCC-8 | | Authors: | Bohm, K, Scrima, A, Fischer, E.S, Gut, H, Thomae, N.H. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
6GMG
 
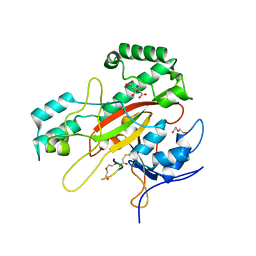 | | Structure of a glutamine donor mimicking inhibitory peptide shaped by the catalytic cleft of microbial transglutaminase | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, PROTEIN-GLUTAMINE GAMMA-GLUTAMYLTRANSFERASE, ... | | Authors: | Schmelz, S, Juettner, N.E, Fuchsbauer, H.L, Scrima, A. | | Deposit date: | 2018-05-25 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a glutamine donor mimicking inhibitory peptide shaped by the catalytic cleft of microbial transglutaminase.
FEBS J., 285, 2018
|
|
6FHP
 
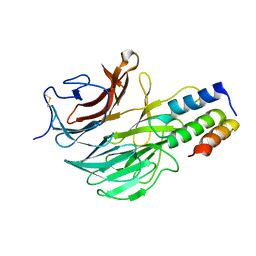 | | DAIP in complex with a C-terminal fragment of thermolysin | | Descriptor: | Dispase autolysis-inducing protein, Thermolysin | | Authors: | Schmelz, S, Fiebig, D, Fuchsbauer, H.L, Blankenfeldt, W, Scrima, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Destructive twisting of neutral metalloproteases: the catalysis mechanism of the Dispase autolysis-inducing protein from Streptomyces mobaraensis DSM 40487.
FEBS J., 285, 2018
|
|
3CP8
 
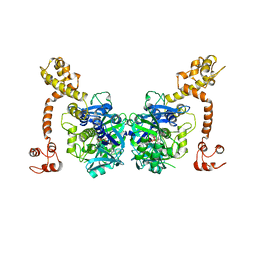 | | Crystal structure of GidA from Chlorobium tepidum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | Authors: | Meyer, S, Scrima, A, Versees, W, Wittinghofer, A. | | Deposit date: | 2008-03-31 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the conserved tRNA-modifying enzyme GidA: implications for its interaction with MnmE and substrate
J.Mol.Biol., 380, 2008
|
|
5OJY
 
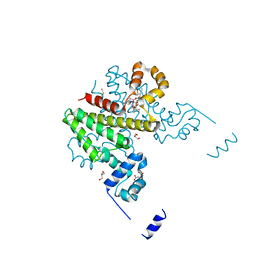 | | Co-complex structure of regulator protein 2 (PamR2) with pamamycin 607 from Streptomyces alboniger | | Descriptor: | CITRIC ACID, GLYCEROL, Pamamycin 607, ... | | Authors: | Schmelz, S, Rebets, Y, Luzhetskyy, A, Scrima, A. | | Deposit date: | 2017-07-24 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Design, development and application of whole-cell based antibiotic-specific biosensor.
Metab. Eng., 47, 2018
|
|
5NPV
 
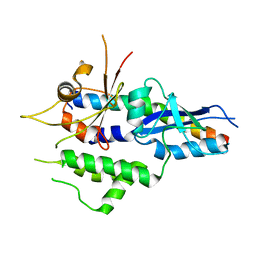 | | Structure of human ATG5-ATG16L1(ATG5BD) complex (I4) | | Descriptor: | Autophagy protein 5, Autophagy-related protein 16-1 | | Authors: | Archna, A, Scrima, A. | | Deposit date: | 2017-04-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification, biochemical characterization and crystallization of the central region of human ATG16L1.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5NPW
 
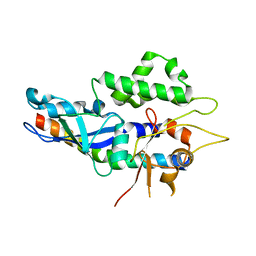 | | Structure of human ATG5-ATG16L1(ATG5BD) complex (C2) | | Descriptor: | Autophagy protein 5, Autophagy-related protein 16-1 | | Authors: | Archna, A, Scrima, A. | | Deposit date: | 2017-04-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification, biochemical characterization and crystallization of the central region of human ATG16L1.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
2HF8
 
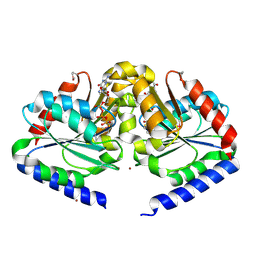 | | Crystal structure of HypB from Methanocaldococcus jannaschii in the triphosphate form, in complex with zinc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Probable hydrogenase nickel incorporation protein hypB, ... | | Authors: | Gasper, R, Scrima, A, Wittinghofer, A. | | Deposit date: | 2006-06-23 | | Release date: | 2006-07-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into HypB, a GTP-binding protein that regulates metal binding.
J.Biol.Chem., 281, 2006
|
|
5OJX
 
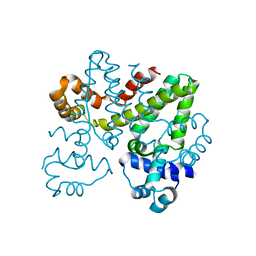 | | Crystal structure of regulator protein 2 (PamR2) from the pamamycin biosynthetic gene cluster of Streptomyces alboniger | | Descriptor: | TetR family transcription regulator | | Authors: | Schmelz, S, Rebets, Y, Luzhetskyy, A, Scrima, A. | | Deposit date: | 2017-07-24 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, development and application of whole-cell based antibiotic-specific biosensor.
Metab. Eng., 47, 2018
|
|
2HF9
 
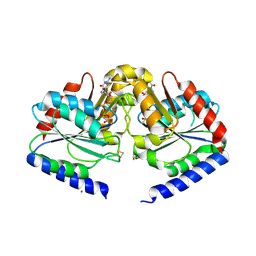 | | Crystal structure of HypB from Methanocaldococcus jannaschii in the triphosphate form | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Probable hydrogenase nickel incorporation protein hypB, ... | | Authors: | Gasper, R, Scrima, A, Wittinghofer, A. | | Deposit date: | 2006-06-23 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into HypB, a GTP-binding protein that regulates metal binding.
J.Biol.Chem., 281, 2006
|
|
5NUV
 
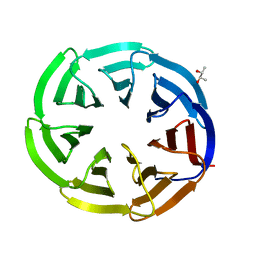 | |
5NTB
 
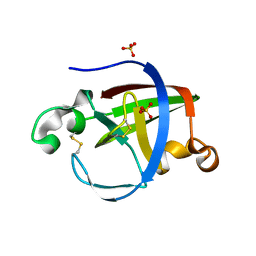 | | Streptomyces papain inhibitor (SPI) | | Descriptor: | Papain inhibitor, SULFATE ION | | Authors: | Schmelz, S, Juettner, N.E, Fuchsbauer, H.L, Scrima, A. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Illuminating structure and acyl donor sites of a physiological transglutaminase substrate from Streptomyces mobaraensis.
Protein Sci., 27, 2018
|
|
4BJT
 
 | | Crystal structure of the Rap1 C-terminal domain (Rap1-RCT) in complex with the Rap1 binding module of Rif1 (Rif1-RBM) | | Descriptor: | 1,2-ETHANEDIOL, DNA-BINDING PROTEIN RAP1, TELOMERE LENGTH REGULATOR PROTEIN RIF1 | | Authors: | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4BJ6
 
 | | Crystal structure Rif2 in complex with the C-terminal domain of Rap1 (Rap1-RCT) | | Descriptor: | DNA-BINDING PROTEIN RAP1, RAP1-INTERACTING FACTOR 2, SULFATE ION | | Authors: | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4A0K
 
 | | STRUCTURE OF DDB1-DDB2-CUL4A-RBX1 BOUND TO A 12 BP ABASIC SITE CONTAINING DNA-DUPLEX | | Descriptor: | 12 BP DNA, 12 BP THF CONTAINING DNA, CULLIN-4A, ... | | Authors: | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (5.93 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0L
 
 | | Structure of DDB1-DDB2-CUL4B-RBX1 bound to a 12 bp abasic site containing DNA-duplex | | Descriptor: | 12 BP DNA DUPLEX, 12 BP THF CONTAINING DNA DUPLEX, CULLIN-4B, ... | | Authors: | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (7.4 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
4BJ5
 
 | | Crystal structure of Rif2 in complex with the C-terminal domain of Rap1 (Rap1-RCT) | | Descriptor: | DNA-BINDING PROTEIN RAP1, PROTEIN RIF2, SULFATE ION | | Authors: | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4BJ1
 
 | | Crystal structure of Saccharomyces cerevisiae RIF2 | | Descriptor: | CHLORIDE ION, PROTEIN RIF2 | | Authors: | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-15 | | Release date: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4BJS
 
 | | Crystal structure of the Rif1 C-terminal domain (Rif1-CTD) from Saccharomyces cerevisiae | | Descriptor: | TELOMERE LENGTH REGULATOR PROTEIN RIF1 | | Authors: | Bunker, R.D, Shi, T, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
5K7H
 
 | | Crystal structure of AibR in complex with the effector molecule isovaleryl coenzyme A | | Descriptor: | CHLORIDE ION, Isovaleryl-coenzyme A, NICKEL (II) ION, ... | | Authors: | Bock, T, Volz, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-26 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The AibR-isovaleryl coenzyme A regulator and its DNA binding site - a model for the regulation of alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Nucleic Acids Res., 45, 2017
|
|
7NBW
 
 | |
6YIZ
 
 | |
2WBL
 
 | | Three-dimensional structure of a binary ROP-PRONE complex | | Descriptor: | RAC-LIKE GTP-BINDING PROTEIN ARAC2, RHO OF PLANTS GUANINE NUCLEOTIDE EXCHANGE FACTOR 8 | | Authors: | Thomas, C, Fricke, I, Weyand, M, Berken, A. | | Deposit date: | 2009-03-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 3D Structure of a Binary Rop-Prone Complex: The Final Intermediate for a Complete Set of Molecular Snapshots of the Ropgef Reaction.
Biol.Chem., 390, 2009
|
|
5K7F
 
 | | Crystal structure of apo AibR | | Descriptor: | ACETATE ION, Transcriptional regulator, TetR family | | Authors: | Bock, T, Volz, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-26 | | Release date: | 2016-12-21 | | Last modified: | 2017-05-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The AibR-isovaleryl coenzyme A regulator and its DNA binding site - a model for the regulation of alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Nucleic Acids Res., 45, 2017
|
|
4B0F
 
 | | Heptameric core complex structure of C4b-binding (C4BP) protein from human | | Descriptor: | C4B-BINDING PROTEIN ALPHA CHAIN, CHLORIDE ION | | Authors: | Schmelz, S, Hofmeyer, T, Kolmar, H, Heinz, D.W. | | Deposit date: | 2012-07-02 | | Release date: | 2013-01-09 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Arranged Sevenfold: Structural Insights Into the C-Terminal Oligomerization Domain of Human C4B-Binding Protein.
J.Mol.Biol., 425, 2013
|
|
