7INS
 
 | | STRUCTURE OF PORCINE INSULIN COCRYSTALLIZED WITH CLUPEINE Z | | Descriptor: | GENERAL PROTAMINE CHAIN, INSULIN (CHAIN A), INSULIN (CHAIN B), ... | | Authors: | Balschmidt, P, Hansen, F.B, Dodson, E, Dodson, G, Korber, F. | | Deposit date: | 1991-09-03 | | Release date: | 1994-01-31 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of porcine insulin cocrystallized with clupeine Z.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
1HBW
 
 | | Solution nmr structure of the dimerization domain of the yeast transcriptional activator Gal4 (residues 50-106) | | Descriptor: | REGULATORY PROTEIN GAL4 | | Authors: | Hidalgo, P, Ansari, A.Z, Schmidt, P, Hare, B, Simkovic, N, Farrell, S, Shin, E.J, Ptashne, M, Wagner, G. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Recruitment of the Transcriptional Machinery Through Gal11P: Structure and Interactions of the GAL4 Dimerization Domain
Genes Dev., 15, 2001
|
|
5FND
 
 | | Dynamic Undocking and the Quasi-Bound State as tools for Drug Design | | Descriptor: | HEAT SHOCK PROTEIN, HSP90-ALPHA, N-(2-AZANYL-6-METHYL-1,3-BENZOTHIAZOL-5-YL)ETHANAMIDE, ... | | Authors: | Ruiz-Carmona, S, Schmidtke, P, Luque, F.J, Baker, L.M, Matassova, N, Davis, B, Roughley, S, Murray, J, Hubbard, R, Barril, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dynamic undocking and the quasi-bound state as tools for drug discovery.
Nat Chem, 9, 2017
|
|
1MHJ
 
 | |
1MHI
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF AN INSULIN DIMER. A STUDY OF THE B9(ASP) MUTANT OF HUMAN INSULIN USING NUCLEAR MAGNETIC RESONANCE DISTANCE GEOMETRY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | INSULIN | | Authors: | Jorgensen, A.M.M, Kristensen, S.M, Led, J.J, Balschmidt, P. | | Deposit date: | 1994-11-30 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of an insulin dimer. A study of the B9(Asp) mutant of human insulin using nuclear magnetic resonance, distance geometry and restrained molecular dynamics.
J.Mol.Biol., 227, 1992
|
|
5FNF
 
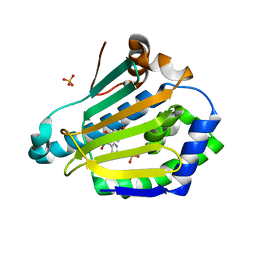 | | Dynamic Undocking and the Quasi-Bound State as tools for Drug Design | | Descriptor: | 4-[(E)-N-oxidanyl-C-pyridin-3-yl-carbonimidoyl]benzene-1,3-diol, HEAT SHOCK PROTEIN, HSP90-ALPHA, ... | | Authors: | Ruiz-Carmona, S, Schmidtke, P, Luque, F.J, Baker, L.M, Matassova, N, Davis, B, Roughley, S, Murray, J, Hubbard, R, Barril, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dynamic undocking and the quasi-bound state as tools for drug discovery.
Nat Chem, 9, 2017
|
|
5FNC
 
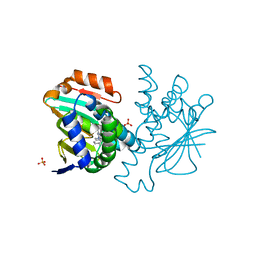 | | Dynamic Undocking and the Quasi-Bound State as tools for Drug Design | | Descriptor: | 6-CHLORO-4-N-[(4-METHYLPHENYL)METHYL]PYRIMIDINE- 2,4-DIAMINE, HEAT SHOCK PROTEIN, HSP90-ALPHA, ... | | Authors: | Ruiz-Carmona, S, Schmidtke, P, Luque, F.J, Baker, L.M, Matassova, N, Davis, B, Roughley, S, Murray, J, Hubbard, R, Barril, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamic undocking and the quasi-bound state as tools for drug discovery.
Nat Chem, 9, 2017
|
|
6MCX
 
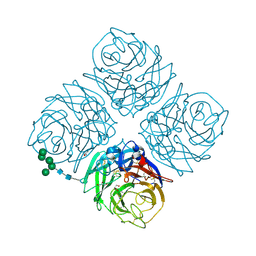 | | INFLUENZA VIRUS NEURAMINIDASE SUBTYPE N9 (TERN) RECOMBINANT HEAD DOMAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Streltsov, V.A, Schmidt, P, McKimm-Breschkin, J. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of an Influenza A virus N9 neuraminidase with a tetrabrachion-domain stalk.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5ZBH
 
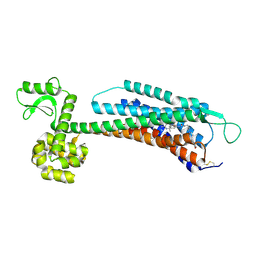 | | The Crystal Structure of Human Neuropeptide Y Y1 Receptor with BMS-193885 | | Descriptor: | Neuropeptide Y receptor type 1,T4 Lysozyme,Neuropeptide Y receptor type 1, dimethyl 4-{3-[({3-[4-(3-methoxyphenyl)piperidin-1-yl]propyl}carbamoyl)amino]phenyl}-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate | | Authors: | Yang, Z, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2018-02-11 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
5ZBQ
 
 | | The Crystal Structure of human neuropeptide Y Y1 receptor with UR-MK299 | | Descriptor: | Neuropeptide Y receptor type 1,T4 Lysozyme, N~2~-(diphenylacetyl)-N-[(4-hydroxyphenyl)methyl]-N~5~-(N'-{[2-(propanoylamino)ethyl]carbamoyl}carbamimidoyl)-D-ornithinamide | | Authors: | Yang, Z, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2018-02-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
6CRD
 
 | | INFLUENZA VIRUS NEURAMINIDASE SUBTYPE N9 (TERN) with tetrabrachion (TB) domain stalk | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Tetrabrachion,Neuraminidase, ... | | Authors: | Streltsov, V.A, Schmidt, P, McKimm-Breschkin, J. | | Deposit date: | 2018-03-16 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of an Influenza A virus N9 neuraminidase with a tetrabrachion-domain stalk.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6D3B
 
 | | INFLUENZA VIRUS NEURAMINIDASE SUBTYPE N9 (TERN) APO FORM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Streltsov, V.A, Schmidt, P, McKimm-Breschkin, J. | | Deposit date: | 2018-04-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of an Influenza A virus N9 neuraminidase with a tetrabrachion-domain stalk.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
2K8I
 
 | | Solution structure of E.Coli SlyD | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Weininger, U, Balbach, J. | | Deposit date: | 2008-09-11 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of SlyD from Escherichia coli: spatial separation of prolyl isomerase and chaperone function.
J.Mol.Biol., 387, 2009
|
|
