4R0B
 
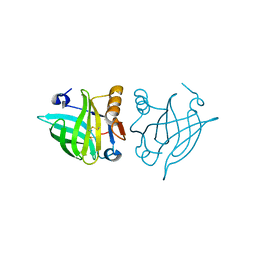 | | Structure of dimeric human glycodelin | | Descriptor: | Glycodelin | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2014-07-30 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The dimeric crystal structure of the human fertility lipocalin glycodelin reveals a protein scaffold for the presentation of complex glycans.
Biochem.J., 466, 2015
|
|
5E9O
 
 | | Spirochaeta thermophila X module - CBM64 - mutant G504A | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Cellulase, glycosyl hydrolase family 5, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2015-10-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for cellulose binding by the type A carbohydrate-binding module 64 of Spirochaeta thermophila.
Proteins, 84, 2016
|
|
5E9P
 
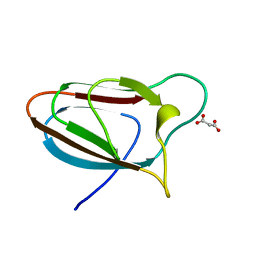 | | Spirochaeta thermophila X module - CBM64 - wildtype | | Descriptor: | Cellulase, glycosyl hydrolase family 5, TPS linker, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2015-10-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for cellulose binding by the type A carbohydrate-binding module 64 of Spirochaeta thermophila.
Proteins, 84, 2016
|
|
6YJW
 
 | | Structure of Fragaria ananassa O-methyltransferase crystallized with PAS polypeptide | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, O-methyltransferase, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Proline/alanine-rich sequence (PAS) polypeptides as an alternative to PEG precipitants for protein crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6YJX
 
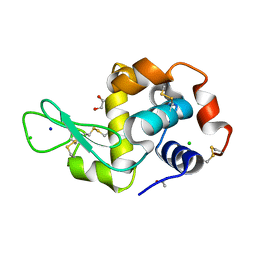 | | Structure of Hen egg-white lysozyme crystallized with PAS polypeptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Proline/alanine-rich sequence (PAS) polypeptides as an alternative to PEG precipitants for protein crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4RUN
 
 | |
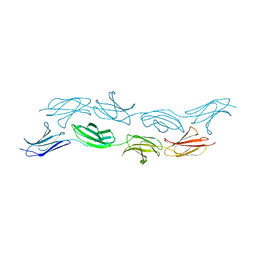
3T1W
 
 | |
4IDE
 
 | | Structure of the Fragaria x ananassa enone oxidoreductase in complex with NADP+ and EDHMF | | Descriptor: | (2E)-2-ethylidene-4-hydroxy-5-methylfuran-3(2H)-one, 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the enzymatic formation of the key strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone
J.Biol.Chem., 288, 2013
|
|
4IDA
 
 | |
4KRX
 
 | | Structure of Aes from E. coli | | Descriptor: | Acetyl esterase, TETRAETHYLENE GLYCOL | | Authors: | Schiefner, A, Gerber, K, Brosig, A, Boos, W. | | Deposit date: | 2013-05-17 | | Release date: | 2013-08-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mutational analyses of Aes, an inhibitor of MalT in Escherichia coli.
Proteins, 82, 2014
|
|
4KRY
 
 | | Structure of Aes from E. coli in covalent complex with PMS | | Descriptor: | Acetyl esterase, IMIDAZOLE, PENTAETHYLENE GLYCOL, ... | | Authors: | Schiefner, A, Gerber, K, Brosig, A, Boos, W. | | Deposit date: | 2013-05-17 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational analyses of Aes, an inhibitor of MalT in Escherichia coli.
Proteins, 82, 2014
|
|
5N48
 
 | |
5N47
 
 | |
1Z6R
 
 | | Crystal structure of Mlc from Escherichia coli | | Descriptor: | Mlc protein, ZINC ION | | Authors: | Schiefner, A, Gerber, K, Seitz, S, Welte, W, Diederichs, K, Boos, W. | | Deposit date: | 2005-03-23 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of Mlc, a global regulator of sugar metabolism in Escherichia coli
J.Biol.Chem., 280, 2005
|
|
5N5U
 
 | |
3LRG
 
 | | Structure of anti-huntingtin VL domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IMIDAZOLE, anti-huntingtin VL domain | | Authors: | Schiefner, A, Chatwell, L, Skerra, A. | | Deposit date: | 2010-02-11 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Disulfide-Free Single-Domain V(L) Intrabody with Blocking Activity towards Huntingtin Reveals a Novel Mode of Epitope Recognition.
J.Mol.Biol., 414, 2011
|
|
3LRH
 
 | |
5N5V
 
 | |
3MBT
 
 | | Structure of monomeric Blc from E. coli | | Descriptor: | Outer membrane lipoprotein blc | | Authors: | Schiefner, A, Skerra, A. | | Deposit date: | 2010-03-26 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analyses reveal a monomeric state of the bacterial lipocalin Blc.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1SW2
 
 | | Crystal structure of ProX from Archeoglobus fulgidus in complex with glycine betaine | | Descriptor: | TRIMETHYL GLYCINE, osmoprotection protein (proX) | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
1SW5
 
 | | Crystal structure of ProX from Archeoglobus fulgidus in the ligand free form | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, osmoprotection protein (proX) | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
1SW4
 
 | | Crystal structure of ProX from Archeoglobus fulgidus in complex with trimethyl ammonium | | Descriptor: | CHLORIDE ION, TETRAMETHYLAMMONIUM ION, ZINC ION, ... | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
1SW1
 
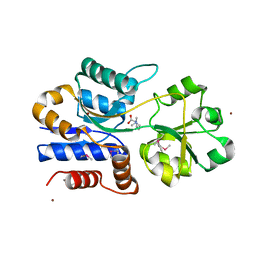 | | Crystal structure of ProX from Archeoglobus fulgidus in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, ZINC ION, osmoprotection protein (proX) | | Authors: | Schiefner, A, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Biol.Chem., 279, 2004
|
|
3GMR
 
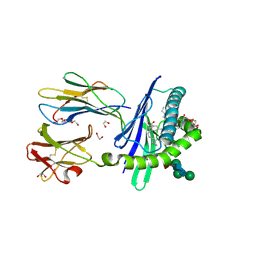 | | Structure of mouse CD1d in complex with C8Ph, different space group | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2 microglobulin, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
3GMM
 
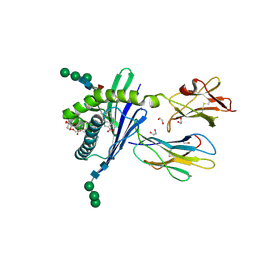 | | Structure of mouse CD1d in complex with C8Ph | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2 microglobulin, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
