2YTA
 
 | | Solution structure of C2H2 type Zinc finger domain 3 in Zinc finger protein 32 | | Descriptor: | ZINC ION, Zinc finger protein 32 | | Authors: | Kasahara, N, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of C2H2 type Zinc finger domain 3 in Zinc finger protein 32
To be Published
|
|
2YTC
 
 | | Solution structure of RNA binding domain in Pre-mRNA-splicing factor RBM22 | | Descriptor: | Pre-mRNA-splicing factor RBM22 | | Authors: | Kasahara, N, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain in Pre-mRNA-splicing factor RBM22
To be Published
|
|
2YTB
 
 | | Solution structure of C2H2 type Zinc finger domain 5 in Zinc finger protein 32 | | Descriptor: | ZINC ION, Zinc finger protein 32 | | Authors: | Kasahara, N, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of C2H2 type Zinc finger domain 5 in Zinc finger protein 32
To be Published
|
|
2YT9
 
 | | Solution structure of C2H2 type Zinc finger domain 345 in Zinc finger protein 278 | | Descriptor: | ZINC ION, zinc finger-containing protein 1 | | Authors: | Kasahara, N, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of C2H2 type Zinc finger domain 345 in Zinc finger protein 278
To be Published
|
|
8GH4
 
 | | Complex of Adam 10 disentegrin cysteine rich domains with human monoclonal antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody heavy chain, Antibody light chain, ... | | Authors: | Nikolov, D.B, Saha, N, Xu, K, Goldgur, Y. | | Deposit date: | 2023-03-09 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Fully human monoclonal antibody targeting activated ADAM10 on colorectal cancer cells.
Biomed Pharmacother, 161, 2023
|
|
5L0Q
 
 | | Crystal structure of the complex between ADAM10 D+C domain and a conformation specific mAb 8C7. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Disintegrin and metalloproteinase domain-containing protein 10, MAGNESIUM ION, ... | | Authors: | Xu, K, Saha, N, Nikolov, D.B. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.759 Å) | | Cite: | An activated form of ADAM10 is tumor selective and regulates cancer stem-like cells and tumor growth.
J.Exp.Med., 213, 2016
|
|
4P91
 
 | | Crystal structure of the nogo-receptor-2 (27-330) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Reticulon-4 receptor-like 2 | | Authors: | Semavina, M, Saha, N, Kolev, M, Goldgur, Y, Giger, R, Himanen, J, Nikolov, D. | | Deposit date: | 2014-04-01 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Nogo-receptor-2.
Protein Sci., 20, 2011
|
|
4P8S
 
 | | Crystal structure of Nogo-receptor-2 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Semavina, M, Saha, N, Kolev, M.V, Giger, R.J, Himanen, J.P, Nikolov, D.B. | | Deposit date: | 2014-04-01 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Nogo-receptor-2.
Protein Sci., 2011
|
|
2AO7
 
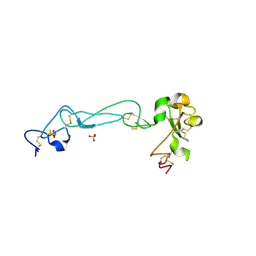 | | Adam10 Disintegrin and cysteine- rich domain | | Descriptor: | ADAM 10, SULFATE ION | | Authors: | Janes, P.W, Saha, N, Barton, W.A, Kolev, M.V, Wimmer-Kleikamp, S.H, Nievergall, E, Blobel, C.P, Himanen, J.-P, Lackmann, M, Nikolov, D.B. | | Deposit date: | 2005-08-12 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans.
Cell(Cambridge,Mass.), 123, 2005
|
|
7NRS
 
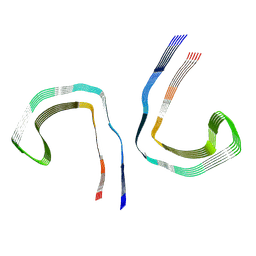 | | Conformation 1 of straight filament from primary age-related tauopathy brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7NRV
 
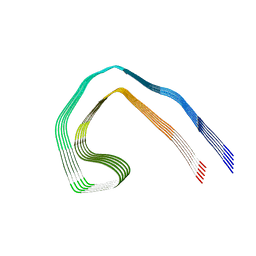 | | Paired helical filament from Alzheimer's disease with PET ligand APN-1607 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7NRQ
 
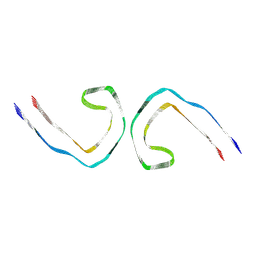 | | Paired helical filament from primary age-related tauopathy brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7NRT
 
 | | Conformation 2 of straight filament from primary age-related tauopathy brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
7NRX
 
 | | Straight filament from Alzheimer's disease with PET ligand APN-1607 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Murzin, A.G, Falcon, B, Epstein, A, Machin, J, Tempest, P, Newell, K.L, Vidal, R, Garringer, H.J, Sahara, N, Higuchi, M, Ghetti, B, Jang, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-03-04 | | Release date: | 2021-03-24 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607.
Acta Neuropathol, 141, 2021
|
|
4I0U
 
 | | Improved structure of Thermotoga maritima CorA at 2.7 A resolution | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nordin, N, Guskov, A, Phua, T, Sahaf, N, Xia, Y, Lu, S.Y, Eshaghi, H, Eshaghi, S. | | Deposit date: | 2012-11-19 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploring the structure and function of Thermotoga maritima CorA reveals the mechanism of gating and ion selectivity in Co2+/Mg2+ transport.
Biochem.J., 451, 2013
|
|
6BDZ
 
 | | ADAM10 Extracellular Domain Bound by the 11G2 Fab | | Descriptor: | 11G2 Fab Heavy Chain, 11G2 Fab Light Chain, CALCIUM ION, ... | | Authors: | Seegar, T.C.M. | | Deposit date: | 2017-10-24 | | Release date: | 2017-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Regulated Proteolysis by the alpha-Secretase ADAM10.
Cell, 171, 2017
|
|
6BE6
 
 | | ADAM10 Extracellular Domain | | Descriptor: | CALCIUM ION, Disintegrin and metalloproteinase domain-containing protein 10, SULFATE ION, ... | | Authors: | Seegar, T.C.M. | | Deposit date: | 2017-10-24 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Regulated Proteolysis by the alpha-Secretase ADAM10.
Cell, 171, 2017
|
|
7S7K
 
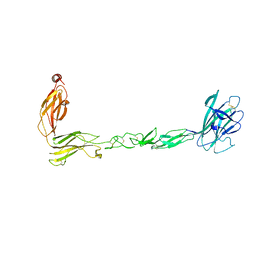 | | Crystal structure of the EphB2 extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-B receptor 2, ... | | Authors: | Xu, Y, Xu, K, Nikolov, D.B. | | Deposit date: | 2021-09-16 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The Ephb2 Receptor Uses Homotypic, Head-to-Tail Interactions within Its Ectodomain as an Autoinhibitory Control Mechanism.
Int J Mol Sci, 22, 2021
|
|
5W4L
 
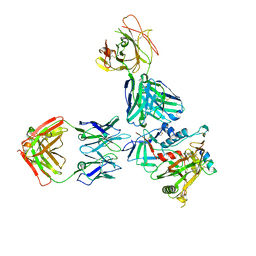 | | Crystal structure of the non-neutralizing and ADCC-potent C11-like antibody N12-i3 in complex with HIV-1 clade A/E gp120, the CD4 mimetic M48U1, and the antibody N5-i5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody N12-i3 Fab heavy chain, Antibody N12-i3 light chain, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Targeting the Late Stage of HIV-1 Entry for Antibody-Dependent Cellular Cytotoxicity: Structural Basis for Env Epitopes in the C11 Region.
Structure, 25, 2017
|
|
